The Characterization of Arabidopsis mterf6 Mutants Reveals a New Role for mTERF6 in Tolerance to Abiotic Stress
Abstract
:1. Introduction
2. Results
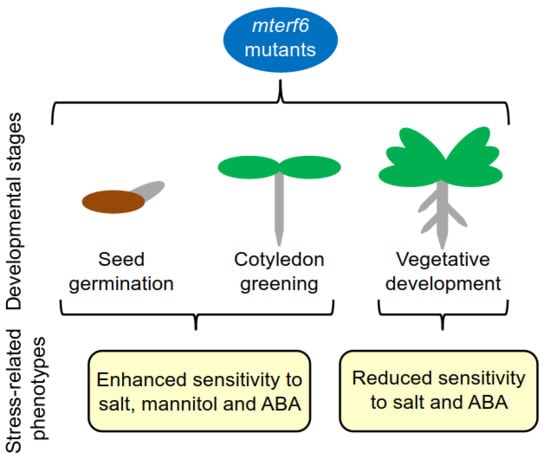
2.1. The mterf6-2 and mterf6-5 Mutants Are Hypersensitive to NaCl and Mannitol
2.2. Knock-Down of mTERF6 Alters the Response to ABA
2.3. The Expression of the mTERF6 Gene Changes in Response to Abiotic Stresses
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Germination and Growth Sensitivity Assays
4.3. Quantitative RT-PCR (qRT-PCR)
4.4. Computational Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| mTERF | mitochondrial transcription termination factor |
| SHOT1 | SUPPRESSOR OF HOT1-4 1 |
| OGE | organellar gene expression |
| SOLDAT10 | SINGLET OXYGEN-LINKED DEATH ACTIVATOR10 |
| MOC1 | mterf-like gene of Chlamydomonas1 |
| ABA | abscisic acid |
| DAS | days after stratification |
| mda1 | mterf defective in Arabidopsis1 |
| RD29A | RESPONSIVE TO DESICCATION29A |
References
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.X.; Crisp, P.A.; Estavillo, G.M.; Pogson, B.J. Chloroplast-to-nucleus communication: Current knowledge, experimental strategies and relationship to drought stress signaling. Plant Signal. Behav. 2010, 5, 1575–1582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leister, D.; Liangsheng, W.; Tatjana, K. Organellar Gene Expression and Acclimation of Plants to Environmental Stress. Front. Plant Sci. 2017, 8, 387. [Google Scholar] [CrossRef] [PubMed]
- Robles, P.; Micol, J.L.; Quesada, V. Arabidopsis MDA1, a nuclear-encoded protein, functions in chloroplast development and abiotic stress responses. PLoS ONE 2012, 7, e42924. [Google Scholar] [CrossRef] [PubMed]
- Robles, P.; Micol, J.L.; Quesada, V. Mutations in the plant-conserved MTERF9 alter chloroplast gene expression, development and tolerance to abiotic stress in Arabidopsis thaliana. Physiol. Plant. 2015, 154, 297–313. [Google Scholar] [CrossRef] [PubMed]
- Linder, T.; Park, C.B.; Asin-Cayuela, J.; Pellegrini, M.; Larsson, N.G.; Falkenberg, M.; Samuelsson, T.; Gustafsson, C.M. A family of putative transcription termination factors shared amongst metazoans and plants. Curr. Genet. 2005, 48, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Leister, D.; Kleine, T. Arabidopsis thaliana mTERF10 and mTERF11, but not mTERF12, are involved in the response to salt stress. Front. Plant Sci. 2017, 8, 1213. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Lee, U.; Small, I.; des Francs-Small, C.C.; Vierling, E. Mutations in an Arabidopsis mitochondrial transcription termination factor-related protein enhance thermotolerance in the absence of the major molecular chaperone HSP101. Plant Cell 2012, 24, 3349–3365. [Google Scholar] [CrossRef] [PubMed]
- Meskauskiene, R.; Würsch, M.; Laloi, C.; Vidi, S.; Coll, N.; Kessler, F.; Baruah, A.; Kim, C.; Apel, K. A mutation in the Arabidopsis mTERF-related plastid protein SOLDAT10 activates retrograde signaling and suppresses 1O2-induced cell death. Plant J. 2009, 60, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Schonfeld, C.; Wobbe, L.; Borgstadt, R.; Kienast, A.; Nixon, P.J.; Kruse, O. The nucleus-encoded protein MOC1 is essential for mitochondrial light acclimation in Chlamydomonas reinhardtii. J. Biol. Chem. 2004, 279, 50366–50374. [Google Scholar] [CrossRef] [PubMed]
- Quesada, V. The roles of mitochondrial transcription termination factors (MTERFs) in plants. Physiol. Plant. 2016, 157, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Roberti, M.; Polosa, P.L.; Bruni, F.; Manzari, C.; Deceglie, S.; Gadaleta, M.N.; Cantatore, P. The MTERF family proteins: Mitochondrial transcription regulators and beyond. Biochim. Biophys. Acta 2009, 1787, 303–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleine, T. Arabidopsis thaliana mTERF proteins: Evolution and functional classification. Front. Plant Sci. 2012, 3, 233. [Google Scholar] [CrossRef] [PubMed]
- Kleine, T.; Leister, D. Emerging functions of mammalian and plant mTERFs. Biochim. Biophys. Acta 2015, 1847, 786–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, Y.W.; Wang, H.J.; Hsieh, M.H.; Hsieh, H.L.; Jauh, G.Y. Arabidopsis mTERF15 is required for mitochondrial nad2 intron 3 splicing and functional complex I activity. PLoS ONE 2014, 9, e112360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammani, K.; Barkan, A. An mTERF domain protein functions in group II intron splicing in maize chloroplasts. Nucleic Acids Res. 2014, 42, 5033–5042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babiychuk, E.; Vandepoele, K.; Wissing, J.; Garcia-Diaz, M.; De Rycke, R.; Akbari, H.; Joubès, J.; Beeckman, T.; Jänsch, L.; Frentzen, M.; et al. Plastid gene expression and plant development require a plastidic protein of the mitochondrial transcription termination factor family. Proc. Natl. Acad. Sci. USA 2011, 108, 6674–6679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quesada, V.; Sarmiento-Mañús, R.; González-Bayón, R.; Hricová, A.; Pérez-Marcos, R.; Graciá-Martínez, E.; Medina-Ruiz, L.; Leyva-Díaz, E.; Ponce, M.R.; Micol, J.L. Arabidopsis RUGOSA2 encodes an mTERF family member required for mitochondrion, chloroplast and leaf development. Plant J. 2011, 68, 738–753. [Google Scholar] [CrossRef] [PubMed]
- Wobbe, L.; Nixon, P.J. The mTERF protein MOC1 terminates mitochondrial DNA transcription in the unicellular green alga Chlamydomonas reinhardtii. Nucleic Acids Res. 2013, 41, 6553–6567. [Google Scholar] [CrossRef] [PubMed]
- Romani, I.; Manavski, N.; Morosetti, A.; Tadini, L.; Maier, S.; Kühn, K.; Ruwe, H.; Schmitz-Linneweber, C.; Wanner, G.; Leister, D.; et al. A Member of the Arabidopsis Mitochondrial Transcription Termination Factor Family Is Required for Maturation of Chloroplast Transfer RNAIle(GAU). Plant Physiol. 2015, 169, 627–646. [Google Scholar] [CrossRef] [PubMed]
- Leister, D.; Kleine, T. Definition of a core module for the nuclear retrograde response to altered organellar gene expression identifies GLK overexpressors as gun mutants. Physiol. Plant. 2016, 157, 297–309. [Google Scholar] [CrossRef] [PubMed]
- Robles, P.; Núñez-Delegido, E.; Ferrández-Ayela, A.; Sarmiento-Mañús, R.; Micol, J.L.; Quesada, V. Arabidopsis mTERF6 is required for leaf patterning. Plant Sci. 2018, 266, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Christmann, A.; Moes, D.; Himmelbach, A.; Yang, Y.; Tang, Y.; Grill, E. Integration of abscisic acid signalling into plant responses. Plant Biol. 2006, 8, 314–325. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Ishitani, M.; Lee, H.; Zhu, J.K. The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress. Plant Cell 2001, 13, 2063–2083. [Google Scholar] [CrossRef] [PubMed]
- González-Guzmán, M.; Apostolova, N.; Bellés, J.M.; Barrero, J.M.; Piqueras, P.; Ponce, M.R.; Micol, J.L.; Serrano, R.; Rodríguez, P. The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 2002, 8, 1833–1846. [Google Scholar] [CrossRef]
- Shkolnik-Inbar, D.; Adler, G.; Bar-Zvi, D. ABI4 downregulates expression of the sodium transporter HKT1;1 in Arabidopsis roots and affects salt tolerance. Plant J. 2013, 73, 993–1005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kilian, J.; Whitehead, D.; Horak, J.; Wanke, D.; Weinl, S.; Batistic, O.; D’Angelo, C.; Bornberg-Bauer, E.; Kudla, J.; Harter, K. The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007, 50, 347–363. [Google Scholar] [CrossRef] [PubMed]
- Zeller, G.; Henz, S.R.; Widmer, C.K.; Sachsenberg, T.; Rätsch, G.; Weigel, D.; Laubinger, S. Stress-induced changes in the Arabidopsis thaliana transcriptome analyzed using whole genome tiling arrays. Plant J. 2009, 58, 1068–1082. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Characterization of the expression of a desiccation-responsive RD29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol. Gen. Genet. 1993, 236, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Cai, M.; Zhang, X.; Li, Y.; Zhang, J.; Zhao, H.; Kong, F.; Zheng, Y.; Qiu, F. Genome-Wide identification, evolution and expression analysis of mTERF gene family in maize. PLoS ONE 2014, 9, e94126. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Zhang, L.; Ullah, A.; Jin, X.; Yang, X.; Zhang, X. Identification of multiple stress responsive genes by sequencing a normalized cDNA library from sea-land cotton (Gossypium barbadense L.). PLoS ONE 2016, 11, e0152927. [Google Scholar] [CrossRef] [PubMed]
- Moschopoulos, A.; Derbyshire, P.; Byrne, M.E. The Arabidopsis organelle-localized glycyl-tRNA synthetase encoded by EMBRYO DEFECTIVE DEVELOPMENT1 is required for organ patterning. J. Exp. Bot. 2012, 63, 5233–5243. [Google Scholar] [CrossRef] [PubMed]

| Genotype | Inhibition of Root Length (%) | |||
|---|---|---|---|---|
| NaCl (mM) | ABA (µM) | |||
| 125 | 150 | 5 | 10 | |
| Col-0 | 64.6 ± 7.2 | 77.2 ± 4.3 | 19.4 ± 7.2 | 29.0 ± 4.5 |
| mterf6-5 | 55.8 ± 6.0 ** | 63.5 ± 4.6 ** | 9.4 ± 13.1 ** | 23.2 ± 14.7 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robles, P.; Navarro-Cartagena, S.; Ferrández-Ayela, A.; Núñez-Delegido, E.; Quesada, V. The Characterization of Arabidopsis mterf6 Mutants Reveals a New Role for mTERF6 in Tolerance to Abiotic Stress. Int. J. Mol. Sci. 2018, 19, 2388. https://doi.org/10.3390/ijms19082388
Robles P, Navarro-Cartagena S, Ferrández-Ayela A, Núñez-Delegido E, Quesada V. The Characterization of Arabidopsis mterf6 Mutants Reveals a New Role for mTERF6 in Tolerance to Abiotic Stress. International Journal of Molecular Sciences. 2018; 19(8):2388. https://doi.org/10.3390/ijms19082388
Chicago/Turabian StyleRobles, Pedro, Sergio Navarro-Cartagena, Almudena Ferrández-Ayela, Eva Núñez-Delegido, and Víctor Quesada. 2018. "The Characterization of Arabidopsis mterf6 Mutants Reveals a New Role for mTERF6 in Tolerance to Abiotic Stress" International Journal of Molecular Sciences 19, no. 8: 2388. https://doi.org/10.3390/ijms19082388
APA StyleRobles, P., Navarro-Cartagena, S., Ferrández-Ayela, A., Núñez-Delegido, E., & Quesada, V. (2018). The Characterization of Arabidopsis mterf6 Mutants Reveals a New Role for mTERF6 in Tolerance to Abiotic Stress. International Journal of Molecular Sciences, 19(8), 2388. https://doi.org/10.3390/ijms19082388