Cytochrome P450 Monooxygenase CYP139 Family Involved in the Synthesis of Secondary Metabolites in 824 Mycobacterial Species
Abstract
:1. Introduction
2. Results and Discussion
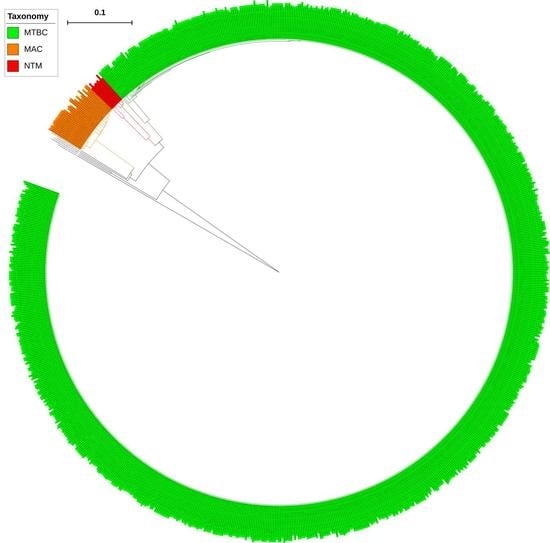
2.1. CYP139 P450s Are Present Only in Certain Mycobacterial Category Species
2.2. CYP139 P450 Family Ranked among Top 10 P450 Families
2.3. CYP139 Family Has Unique Amino Acid Patterns at CXG Motif
2.4. Most CYP139A P450s Are Part of Secondary Metabolite Biosynthetic Gene Clusters
2.5. CYP139A P450s Involved in the Synthesis of Secondary Metabolites in Mycobacterial Species
3. Materials and Methods
3.1. Mycobacterial Species and Genome Databases
3.2. Genome Data Mining and Annotation of CYP139 P450s
3.3. Phylogenetic Analysis of CYP139A P450s
3.4. Analysis of Homology and Amino Acid Conservation
3.5. Generation of EXXR and CXG Sequence Logo
3.6. Identification of CYP139 P450 Secondary Metabolite BGCs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization (WHO). Global Tuberculosis Report 2018. Available online: https://www.who.int/tb/publications/global_report/en/ (accessed on 22 March 2019).
- Mortality and Causes of Death in South Africa, 2016: Findings from Death Notification; Statistics South Africa: Pretoria, South Africa, 2018. Available online: http://www.statssa.gov.za/publications/P03093/P030932016.pdf (accessed on 22 March 2019).
- Cole, S.T.; Brosch, R.; Parkhill, J.; Garnier, T.; Churcher, C.; Harris, D.; Gordon, S.V.; Eiglmeier, K.; Gas, S.; Barry, C.E.; et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998, 393, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.R. Cytochrome P450 diversity in the tree of life. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 141–154. [Google Scholar] [CrossRef]
- Ortiz de Montellano, P.R. Potential drug targets in the Mycobacterium tuberculosis cytochrome P450 system. J. Inorg. Biochem. 2018, 180, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Bellamine, A.; Mangla, A.T.; Dennis, A.L.; Nes, W.D.; Waterman, M.R. Structural requirements for substrate recognition of Mycobacterium tuberculosis 14 alpha-demethylase: Implications for sterol biosynthesis. J. Lipid Res. 2001, 42, 128–136. [Google Scholar] [PubMed]
- Bellamine, A.; Mangla, A.T.; Nes, W.D.; Waterman, M.R. Characterization and catalytic properties of the sterol 14alpha-demethylase from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 1999, 96, 8937–8942. [Google Scholar] [CrossRef] [PubMed]
- McLean, K.J.; Warman, A.J.; Seward, H.E.; Marshall, K.R.; Girvan, H.M.; Cheesman, M.R.; Waterman, M.R.; Munro, A.W. Biophysical characterization of the sterol demethylase P450 from Mycobacterium tuberculosis, its cognate ferredoxin, and their interactions. Biochemistry 2006, 45, 8427–8443. [Google Scholar] [CrossRef]
- Belin, P.; Le Du, M.H.; Fielding, A.; Lequin, O.; Jacquet, M.; Charbonnier, J.B.; Lecoq, A.; Thai, R.; Courcon, M.; Masson, C.; et al. Identification and structural basis of the reaction catalyzed by CYP121, an essential cytochrome P450 in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2009, 106, 7426–7431. [Google Scholar] [CrossRef]
- Johnston, J.B.; Ouellet, H.; Ortiz de Montellano, P.R. Functional redundancy of steroid C26-monooxygenase activity in Mycobacterium tuberculosis revealed by biochemical and genetic analyses. J. Biol. Chem. 2010, 285, 36352–36360. [Google Scholar] [CrossRef] [PubMed]
- Driscoll, M.D.; McLean, K.J.; Levy, C.; Mast, N.; Pikuleva, I.A.; Lafite, P.; Rigby, S.E.; Leys, D.; Munro, A.W. Structural and biochemical characterization of Mycobacterium tuberculosis CYP142: Evidence for multiple cholesterol 27-hydroxylase activities in a human pathogen. J. Biol. Chem. 2010, 285, 38270–38282. [Google Scholar] [CrossRef] [PubMed]
- Johnston, J.B.; Kells, P.M.; Podust, L.M.; Ortiz de Montellano, P.R. Biochemical and structural characterization of CYP124: A methyl-branched lipid omega-hydroxylase from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2009, 106, 20687–20692. [Google Scholar] [CrossRef] [PubMed]
- Johnston, J.B.; Singh, A.A.; Clary, A.A.; Chen, C.K.; Hayes, P.Y.; Chow, S.; De Voss, J.J.; Ortiz de Montellano, P.R. Substrate analog studies of the omega-regiospecificity of Mycobacterium tuberculosis cholesterol metabolizing cytochrome P450 enzymes CYP124A1, CYP125A1 and CYP142A1. Bioorg. Med. Chem. 2012, 20, 4064–4081. [Google Scholar] [CrossRef] [PubMed]
- Sogi, K.M.; Holsclaw, C.M.; Fragiadakis, G.K.; Nomura, D.K.; Leary, J.A.; Bertozzi, C.R. Biosynthesis and regulation of sulfomenaquinone, a metabolite associated with virulence in Mycobacterium tuberculosis. ACS Infect. Dis. 2016, 2, 800–806. [Google Scholar] [CrossRef]
- McLean, K.J.; Munro, A.W. Structural biology and biochemistry of cytochrome P450 systems in Mycobacterium tuberculosis. Drug Metab. Rev. 2008, 40, 427–446. [Google Scholar] [CrossRef] [PubMed]
- Ouellet, H.; Johnston, J.B.; Ortiz de Montellano, P.R. The Mycobacterium tuberculosis cytochrome P450 system. Arch Biochem. Biophys. 2010, 493, 82–95. [Google Scholar] [CrossRef] [PubMed]
- Griffin, J.E.; Gawronski, J.D.; Dejesus, M.A.; Ioerger, T.R.; Akerley, B.J.; Sassetti, C.M. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 2011, 7, e1002251. [Google Scholar] [CrossRef]
- Sassetti, C.M.; Boyd, D.H.; Rubin, E.J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003, 48, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; de Bruijn, I.; Chooi, Y.H.; Claesen, J.; Coates, R.C.; et al. Minimum information about a biosybnthetic gene cluster. Nat. Chem. Biol. 2015, 11, 625–631. [Google Scholar] [CrossRef]
- Greule, A.; Stok, J.E.; De Voss, J.J.; Cryle, M.J. Unrivalled diversity: The many roles and reactions of bacterial cytochromes P450 in secondary metabolism. Nat. Prod. Rep. 2018, 35, 757–791. [Google Scholar] [CrossRef] [PubMed]
- Mthethwa, B.C.; Chen, W.; Ngwenya, M.L.; Kappo, A.P.; Syed, P.R.; Karpoormath, R.; Yu, J.H.; Nelson, D.R.; Syed, K. Comparative analyses of cytochrome P450s and those associated with secondary metabolism in Bacillus species. Int. J. Mol. Sci. 2018, 19, 3623. [Google Scholar] [CrossRef]
- Senate, L.M.; Tjatji, M.P.; Pillay, K.; Chen, W.; Zondo, N.M.; Syed, P.R.; Mnguni, F.C.; Chiliza, Z.E.; Bamal, H.D.; Karpoormath, R.; et al. Similarities, variations, and evolution of cytochrome P450s in Streptomyces versus Mycobacterium. Sci. Rep. 2019, 9, 3962. [Google Scholar] [CrossRef]
- Parvez, M.; Qhanya, L.B.; Mthakathi, N.T.; Kgosiemang, I.K.; Bamal, H.D.; Pagadala, N.S.; Xie, T.; Yang, H.; Chen, H.; Theron, C.W.; et al. Molecular evolutionary dynamics of cytochrome P450 monooxygenases across kingdoms: Special focus on mycobacterial P450s. Sci. Rep. 2016, 6, 33099. [Google Scholar] [CrossRef] [PubMed]
- Jawallapersand, P.; Mashele, S.S.; Kovacic, L.; Stojan, J.; Komel, R.; Pakala, S.B.; Krasevec, N.; Syed, K. Cytochrome P450 monooxygenase CYP53 family in fungi: Comparative structural and evolutionary analysis and its role as a common alternative anti-fungal drug target. PLoS ONE 2014, 9, e107209. [Google Scholar] [CrossRef] [PubMed]
- Bamal, H.D.; Chen, W.; Mashele, S.S.; Nelson, D.R.; Kappo, A.P.; Mosa, R.A.; Yu, J.H.; Tuszynski, J.A.; Syed, K. Comparative analyses and structural insights of the novel cytochrome P450 fusion protein family CYP5619 in Oomycetes. Sci. Rep. 2018, 8, 6597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pei, J.; Kim, B.H.; Grishin, N.V. PROMALS3D: A tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008, 36, 2295–2300. [Google Scholar] [CrossRef]
- Pei, J.; Grishin, N.V. AL2CO: Calculation of positional conservation in a protein sequence alignment. Bioinformatics (Oxf. Engl.) 2001, 17, 700–712. [Google Scholar] [CrossRef]
- Syed, K.; Mashele, S.S. Comparative analysis of P450 signature motifs EXXR and CXG in the large and diverse kingdom of fungi: Identification of evolutionarily conserved amino acid patterns characteristic of P450 family. PLoS ONE 2014, 9, e95616. [Google Scholar] [CrossRef]
- Sello, M.M.; Jafta, N.; Nelson, D.R.; Chen, W.; Yu, J.H.; Parvez, M.; Kgosiemang, I.K.; Monyaki, R.; Raselemane, S.C.; Qhanya, L.B.; et al. Diversity and evolution of cytochrome P450 monooxygenases in Oomycetes. Sci. Rep. 2015, 5, 11572. [Google Scholar] [CrossRef]
- Parvez, A.; Giri, S.; Giri, G.R.; Kumari, M.; Bisht, R.; Saxena, P. Novel Type III polyketide synthases biosynthesize methylated polyketides in Mycobacterium marinum. Sci. Rep. 2018, 8, 6529. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Komaki, H.; Ichikawa, N.; Hosoyama, A.; Sato, S.; Igarashi, Y. Biosynthesis of akaeolide and lorneic acids and annotation of type I polyketide synthase gene clusters in the genome of Streptomyces sp. NPS554. Mar. Drugs 2015, 13, 581–596. [Google Scholar] [CrossRef]
- Ueda, J.Y.; Hashimoto, J.; Yamamura, H.; Hayakawa, M.; Takagi, M.; Shin-ya, K. A new 16-membered tetraene macrolide JBIR-100 from a newly identified Streptomyces species. J. Antibiot. 2010, 63, 627–629. [Google Scholar] [CrossRef]
- Huss, M.; Wieczorek, H. Inhibitors of V-ATPases: Old and new players. J. Exp. Biol. 2009, 212, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Iwata, F.; Sato, S.; Mukai, T.; Yamada, S.; Takeo, J.; Abe, A.; Okita, T.; Kawahara, H. Lorneic acids, trialkyl-substituted aromatic acids from a marine-derived actinomycete. J. Nat. Prod. 2009, 72, 2046–2048. [Google Scholar] [CrossRef] [PubMed]
- Salituro, G.M.; Zink, D.L.; Dahl, A.; Nielsen, J.; Wu, E.; Huang, L.; Kastner, C.; Dumont, F.J. Meridamycin: A novel nonimmunosuppressive FKBP12 ligand from Streptomyces hygroscopicus. Tetrahedron Lett. 1995, 36, 997–1000. [Google Scholar] [CrossRef]
- Aghdasi, B.; Ye, K.; Resnick, A.; Huang, A.; Ha, H.C.; Guo, X.; Dawson, T.M.; Dawson, V.L.; Snyder, S.H. FKBP12, the 12-kDa FK506-binding protein, is a physiologic regulator of the cell cycle. Proc. Natl. Acad. Sci. USA 2001, 98, 2425–2430. [Google Scholar] [CrossRef] [Green Version]
- Wawrocki, S.; Druszczynska, M. Inflammasomes in Mycobacterium tuberculosis-Driven Immunity. Can. J. Infect. Dis. Med Microbiol. J. Can. Mal. Infect. Microbiol. Med 2017, 2017, 2309478. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.P.; Alonso, S.; Rand, L.; Dick, T.; Pethe, K. The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2008, 105, 11945–11950. [Google Scholar] [CrossRef]
- Katsnelson, M.A.; Rucker, L.G.; Russo, H.M.; Dubyak, G.R. K+ efflux agonists induce NLRP3 inflammasome activation independently of Ca2+ signaling. J. Immunol. (Baltim. Md. 1950) 2015, 194, 3937–3952. [Google Scholar] [CrossRef] [PubMed]
- Sarfo, F.S.; Phillips, R.; Wansbrough-Jones, M.; Simmonds, R.E. Recent advances: Role of mycolactone in the pathogenesis and monitoring of Mycobacterium ulcerans infection/Buruli ulcer disease. Cell. Microbiol. 2016, 18, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, H.; Degnes, K.F.; Dikiy, A.; Fjaervik, E.; Klinkenberg, G.; Zotchev, S.B. Insights into the evolution of macrolactam biosynthesis through cloning and comparative analysis of the biosynthetic gene cluster for a novel macrocyclic lactam, ML-449. Appl. Environ. Microbiol. 2010, 76, 283–293. [Google Scholar] [CrossRef]
- Brautaset, T.; Sekurova, O.N.; Sletta, H.; Ellingsen, T.E.; StrLm, A.R.; Valla, S.; Zotchev, S.B. Biosynthesis of the polyene antifungal antibiotic nystatin in Streptomyces noursei ATCC 11455: Analysis of the gene cluster and deduction of the biosynthetic pathway. Chem. Biol. 2000, 7, 395–403. [Google Scholar] [CrossRef]
- Julien, B.; Tian, Z.Q.; Reid, R.; Reeves, C.D. Analysis of the ambruticin and jerangolid gene clusters of Sorangium cellulosum reveals unusual mechanisms of polyketide biosynthesis. Chem. Biol. 2006, 13, 1277–1286. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kong, L.; Shen, J.; Wang, Q.; Liu, Q.; Yang, W.; Deng, Z.; You, D. Characterization of the positive SARP family regulator PieR for improving piericidin A1 production in Streptomyces piomogeues var. Hangzhouwanensis. Synth. Syst. Biotechnol. 2019, 4, 16–24. [Google Scholar] [CrossRef]
- Distler, J.; Ebert, A.; Mansouri, K.; Pissowotzki, K.; Stockmann, M.; Piepersberg, W. Gene cluster for streptomycin biosynthesis in Streptomyces griseus: Nucleotide sequence of three genes and analysis of transcriptional activity. Nucleic Acids Res. 1987, 15, 8041–8056. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhou, X.; Dong, H.; Tu, G.; Wang, M.; Wang, B.; Deng, Z. A complete gene cluster from Streptomyces nanchangensis NS3226 encoding biosynthesis of the polyether ionophore nanchangmycin. Chem. Biol. 2003, 10, 431–441. [Google Scholar] [CrossRef]
- Rausch, K.; Hackett, B.A.; Weinbren, N.L.; Reeder, S.M.; Sadovsky, Y.; Hunter, C.A.; Schultz, D.C.; Coyne, C.B.; Cherry, S. Screening bioactives reveals nanchangmycin as a broad spectrum antiviral active against Zika virus. Cell Rep. 2017, 18, 804–815. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Lin, X.; Zhou, X.; Deng, Z.; Cane, D.E. Mechanism of thioesterase-catalyzed chain release in the biosynthesis of the polyether antibiotic nanchangmycin. Chem. Biol. 2008, 15, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Cassinelli, G.; Grein, A.; Orezzi, P.; Pennella, P.; Sanfilippo, A. New antibiotics produced by Streptoverticillium orinoci, n. sp. Arch. Fur Mikrobiol. 1967, 55, 358–368. [Google Scholar] [CrossRef]
- Elnakady, Y.A.; Chatterjee, I.; Bischoff, M.; Rohde, M.; Josten, M.; Sahl, H.G.; Herrmann, M.; Muller, R. Investigations to the antibacterial mechanism of action of kendomycin. PLoS ONE 2016, 11, e0146165. [Google Scholar] [CrossRef]
- McAlpine, J.B.; Bachmann, B.O.; Piraee, M.; Tremblay, S.; Alarco, A.M.; Zazopoulos, E.; Farnet, C.M. Microbial genomics as a guide to drug discovery and structural elucidation: ECO-02301, a novel antifungal agent, as an example. J. Nat. Prod. 2005, 68, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Li, S.; Niu, S.; Ma, L.; Zhang, G.; Zhang, H.; Zhang, G.; Ju, J.; Zhang, C. Characterization of tiacumicin B biosynthetic gene cluster affording diversified tiacumicin analogues and revealing a tailoring dihalogenase. J. Am. Chem. Soc. 2011, 133, 1092–1105. [Google Scholar] [CrossRef]
- Glaus, F.; Altmann, K.H. Total synthesis of the tiacumicin B (lipiarmycin A3/fidaxomicin) aglycone. Angew. Chem. (Int. Ed. Engl.) 2015, 54, 1937–1940. [Google Scholar] [CrossRef]
- Salomon, A.R.; Voehringer, D.W.; Herzenberg, L.A.; Khosla, C. Apoptolidin, a selective cytotoxic agent, is an inhibitor of F0F1-ATPase. Chem. Biol. 2001, 8, 71–80. [Google Scholar] [CrossRef] [Green Version]
- Wender, P.A.; Sukopp, M.; Longcore, K. Apoptolidins B and C: Isolation, structure determination, and biological activity. Org. Lett. 2005, 7, 3025–3028. [Google Scholar] [CrossRef]
- Kim, J.W.; Adachi, H.; Shin-ya, K.; Hayakawa, Y.; Seto, H. Apoptolidin, a new apoptosis inducer in transformed cells from Nocardiopsis sp. J. Antibiot. 1997, 50, 628–630. [Google Scholar] [CrossRef]
- Sadaka, C.; Ellsworth, E.; Hansen, P.R.; Ewin, R.; Damborg, P.; Watts, J.L. Review on abyssomicins: Inhibitors of the chorismate pathway and folate biosynthesis. Molecules (Basel Switz.) 2018, 23, 1371. [Google Scholar] [CrossRef]
- Skellam, E.J.; Stewart, A.K.; Strangman, W.K.; Wright, J.L. Identification of micromonolactam, a new polyene macrocyclic lactam from two marine Micromonospora strains using chemical and molecular methods: Clarification of the biosynthetic pathway from a glutamate starter unit. J. Antibiot. 2013, 66, 431–441. [Google Scholar] [CrossRef]
- Isogai, A.; Sakuda, S.; Matsumoto, S.; Ogura, M.; Furihata, K.; Seto, H.; Suzuki, A. The structure of leucanicidin, a novel insecticidal macrolide produced by Streptomyces halstedii. Agric. Biol. Chem. 1984, 48, 1379–1381. [Google Scholar] [CrossRef]
- Symersky, J.; Osowski, D.; Walters, D.E.; Mueller, D.M. Oligomycin frames a common drug-binding site in the ATP synthase. Proc. Natl. Acad. Sci. USA 2012, 109, 13961–13965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niggemann, J.; Bedorf, N.; Flörke, U.; Steinmetz, H.; Gerth, K.; Reichenbach, H.; Höfle, G. Spirangien A and B, highly cytotoxic and antifungal spiroketals from the myxobacterium Sorangium cellulosum: Isolation, structure elucidation and chemical modifications. Eur. J. Org. Chem. 2005, 2005, 5013–5018. [Google Scholar] [CrossRef]
- Hasenbohler, A.; Kneifel, H.; Konig, W.A.; Zahner, H.; Zeiler, H.J. Metabolic products of microorganisms. 134. Stenothricin, a new inhibitor of the bacterial cell wall synthesis (author’s transl). Arch. Microbiol. 1974, 99, 307–321. [Google Scholar]
- Habibi, D.; Ogloff, N.; Jalili, R.B.; Yost, A.; Weng, A.P.; Ghahary, A.; Ong, C.J. Borrelidin, a small molecule nitrile-containing macrolide inhibitor of threonyl-tRNA synthetase, is a potent inducer of apoptosis in acute lymphoblastic leukemia. Investig. New Drugs 2012, 30, 1361–1370. [Google Scholar] [CrossRef]
- Olano, C.; Wilkinson, B.; Sanchez, C.; Moss, S.J.; Sheridan, R.; Math, V.; Weston, A.J.; Brana, A.F.; Martin, C.J.; Oliynyk, M.; et al. Biosynthesis of the angiogenesis inhibitor borrelidin by Streptomyces parvulus Tu4055: Cluster analysis and assignment of functions. Chem. Biol. 2004, 11, 87–97. [Google Scholar] [PubMed]
- Kataoka, T.; Yamada, A.; Bando, M.; Honma, T.; Mizoue, K.; Nagai, K. FD-891, a structural analogue of concanamycin A that does not affect vacuolar acidification or perforin activity, yet potently prevents cytotoxic T lymphocyte-mediated cytotoxicity through the blockage of conjugate formation. Immunology 2000, 100, 170–177. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, J.; Zhu, J.; Chen, S.; Bai, L.; Zhou, X.; Wu, H.; Deng, Z. Incomplete beta-ketone processing as a mechanism for polyene structural variation in the FR-008/candicidin complex. Chem. Biol. 2008, 15, 629–638. [Google Scholar] [CrossRef] [PubMed]
- Vetcher, L.; Menzella, H.G.; Kudo, T.; Motoyama, T.; Katz, L. The antifungal polyketide ambruticin targets the HOG pathway. Antimicrob. Agents Chemother. 2007, 51, 3734–3736. [Google Scholar] [CrossRef]
- Chen, I.A.; Chu, K.; Palaniappan, K.; Pillay, M.; Ratner, A.; Huang, J.; Huntemann, M.; Varghese, N.; White, J.R.; Seshadri, R.; et al. IMG/M v.5.0: An integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 2019, 47, D666–D677. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.R. Cytochrome P450 nomenclature. Methods Mol. Biol. (Clifton N.J.) 1998, 107, 15–24. [Google Scholar]
- Nelson, D.R. Cytochrome P450 nomenclature, 2004. Methods Mol. Biol. (Clifton N.J.) 2006, 320, 1–10. [Google Scholar]
- Nelson, D.R.; Kamataki, T.; Waxman, D.J.; Guengerich, F.P.; Estabrook, R.W.; Feyereisen, R.; Gonzalez, F.J.; Coon, M.J.; Gunsalus, I.C.; Gotoh, O.; et al. The P450 superfamily: Update on new sequences, gene mapping, accession numbers, early trivial names of enzymes, and nomenclature. DNA Cell Biol. 1993, 12, 1–51. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Boc, A.; Diallo, A.B.; Makarenkov, V. T-REX: A web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 2012, 40, W573–W579. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- McWilliam, H.; Li, W.; Uludag, M.; Squizzato, S.; Park, Y.M.; Buso, N.; Cowley, A.P.; Lopez, R. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 2013, 41, W597–W600. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Pascal Andreu, V.; de Los Santos, E.L.C.; Del Carratore, F.; Lee, S.Y.; Medema, M.H.; Weber, T. The antiSMASH database version 2: A comprehensive resource on secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2019, 47, D625–D630. [Google Scholar] [CrossRef] [PubMed]





| P450 Family | Number of Member P450s | Kingdom | PROMALS3D Conservation Index | Rank (Highest to Lowest Conservation) | ||||
|---|---|---|---|---|---|---|---|---|
| 5 | 6 | 7 | 8 | 9 | ||||
| CYP141 | 29 | Bacteria | 0 | 0 | 0 | 0 | 389 | 1 |
| CYP51 | 50 | Bacteria | 11 | 102 | 0 | 0 | 264 | 2 |
| CYP137 | 38 | Bacteria | 145 | 0 | 0 | 0 | 251 | 3 |
| CYP121 | 34 | Bacteria | 0 | 0 | 0 | 0 | 233 | 4 |
| CYP132 | 39 | Bacteria | 175 | 0 | 0 | 0 | 217 | 5 |
| CYP5619 | 23 | Stramenopila (oomycetes) | 118 | 38 | 170 | 0 | 199 | 6 |
| CYP124 | 71 | Bacteria | 52 | 35 | 59 | 0 | 170 | 7 |
| CYP139 | 894 | Bacteria | 0 | 127 | 0 | 0 | 165 | 8 (formerly 12) |
| CYP188 | 67 | Bacteria | 62 | 0 | 100 | 0 | 141 | 9 |
| CYP123 | 74 | Bacteria | 62 | 0 | 82 | 0 | 137 | 10 |
| Gene Cluster | Function | Reference |
|---|---|---|
| ML-449 | Macrolactam antifungal-antibiotic production. | [41] |
| MAR/MAP | Synthesis of methylated alkyl-resorcinol and methylated acyl-phloroglucinol products found to be part of cell envelope in M. marinum. | [30] |
| Nystatin | Polyene antifungal antibiotic. | [42] |
| Jerangolid | Antifungal polyketide. | [43] |
| Piericidin A1 | A member of α-pyridone antibiotics, exhibits various biological activities such as antimicrobial, antifungal, and antitumour properties and possesses potent respiration-inhibitory activity against insects owing to its competitive binding capacity to mitochondrial complex I. | [44] |
| Streptomycin | Antibiotic used to treat bacterial infections, including tuberculosis. | [45] |
| Nanchangmycin | A polyether ionophore antibiotic produced by Streptomyces nanchangensis NS3226 that has insecticidal and in vitro antibacterial properties. Nanchangmycin exhibits antiviral properties against the Zika virus. | [46,47,48] |
| Neoaureothin | Neoaureothin is an unusual chain-extended analog of aureothin. It was first reported as a co-metabolite of neoantimycin in Streptomyces orinoci. It has been reported to have anti-HIV and antifungal activity. | [49] |
| Akaeolide | A carbocyclic polyketide with moderate antimicrobial activity and cytotoxicity to rat fibroblasts. | [31] |
| Kendomycin | Macrolide antibiotic with antibacterial activity. | [50] |
| ECO-02301 | Antifungal agent. | [51] |
| Tiacumicin B | Macrolide antibiotic, which is used for the treatment of Clostridium difficile infections. | [52,53] |
| Apoptolidin | Macrolide antibiotic well known as apoptosis inducer and inhibitor of F0F1-ATPase. It is a promising new therapeutic lead that exhibits remarkable selectivity against cancer cells relative to normal cells. | [54,55,56] |
| Abyssomicin | A novel spirotetronate polyketide Class I antimicrobial. The biological activity of abyssomicins includes their antimicrobial activity against Gram-positive bacteria and mycobacteria, antitumour properties, latent HIV reactivator, anti-HIV and HIV replication inducer properties | [57] |
| JBIR-100 | A new 16-membered tetraene macrolide from the Streptomyces species. Its structure is identical to TS155-2, which is an inhibitor of the thrombin-induced calcium influx. It exhibits cytotoxic and V-ATPases inhibition activities. V-ATPases are ubiquitous proton pumps present in the endomembrane system of all eukaryotic cells and in the plasma membranes of many animal cells that have been correlated with an increasing number of diseases such as osteopetrosis, male infertility and renal acidosis. | [32,33] |
| Micromonolactam | A new polyene macrolactam antibiotic | [58] |
| Lorneic acid A | It has a fatty acid-like structure in which a benzene ring is embedded. It inhibits phosphodiesterases (PDE) with selectivity toward PDE5, thus, blocking the degradation of cGMP and having a possible linkage to pulmonary hypertension | [34] |
| Leucanicidin | A potent nematocide and insecticide macrolide | [59] |
| Oligomycin | A natural antibiotic that inhibits mitochondrial ATP synthase, thus affecting the electron transport chain. | [60] |
| Spirangien | Highly cytotoxic and antifungal spiroketal | [61] |
| Stenothricin | A peptide antibiotic inhibiting bacterial cell wall synthesis | [62] |
| Borrelidin | A small molecule nitrile-containing macrolide, which is an inhibitor of bacterial and eukaryal threonyl-tRNA synthetase. It exhibits among others antibacterial and anti-angiogenesis activities, suppresses growth and induces apoptosis in malignant acute lymphoblastic leukemia cells. | [63,64] |
| FD-891 | Profoundly blocked both perforin- and FasL-dependent cytotoxicity by cytotoxic T lymphocytes—immunosuppressive. | [65] |
| FR-008 | Macrolide antibiotic with antifungal activity. | [66] |
| Meridamycin | A 27-membered macrolide that acts as non-immunosuppressive FK506-binding proteins (FKBP12) ligand. | [35] |
| Ambruticin | Antifungal polyketide | [67] |
| Nigericin | Nigericin acts as an H+, K+, Pb2+ ionophore. Most commonly it is an antiporter of H+ and K+. In the past nigericin was used as an antibiotic active against Gram-positive bacteria. It inhibits Golgi functions in eukaryotic cells. Its ability to induce K+ efflux also makes it a potent activator of the NLRP3 inflammasome. | [37,38,39] |
| Mycolactone | Lipid-like toxin with cytotoxic, immunosuppressive and tissue necrosis activity. It plays a key role in the development of Buruli ulcer by M. ulcerans. | [40] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Syed, P.R.; Chen, W.; Nelson, D.R.; Kappo, A.P.; Yu, J.-H.; Karpoormath, R.; Syed, K. Cytochrome P450 Monooxygenase CYP139 Family Involved in the Synthesis of Secondary Metabolites in 824 Mycobacterial Species. Int. J. Mol. Sci. 2019, 20, 2690. https://doi.org/10.3390/ijms20112690
Syed PR, Chen W, Nelson DR, Kappo AP, Yu J-H, Karpoormath R, Syed K. Cytochrome P450 Monooxygenase CYP139 Family Involved in the Synthesis of Secondary Metabolites in 824 Mycobacterial Species. International Journal of Molecular Sciences. 2019; 20(11):2690. https://doi.org/10.3390/ijms20112690
Chicago/Turabian StyleSyed, Puleng Rosinah, Wanping Chen, David R. Nelson, Abidemi Paul Kappo, Jae-Hyuk Yu, Rajshekhar Karpoormath, and Khajamohiddin Syed. 2019. "Cytochrome P450 Monooxygenase CYP139 Family Involved in the Synthesis of Secondary Metabolites in 824 Mycobacterial Species" International Journal of Molecular Sciences 20, no. 11: 2690. https://doi.org/10.3390/ijms20112690
APA StyleSyed, P. R., Chen, W., Nelson, D. R., Kappo, A. P., Yu, J. -H., Karpoormath, R., & Syed, K. (2019). Cytochrome P450 Monooxygenase CYP139 Family Involved in the Synthesis of Secondary Metabolites in 824 Mycobacterial Species. International Journal of Molecular Sciences, 20(11), 2690. https://doi.org/10.3390/ijms20112690