mRNA Engineering for the Efficient Chaperone-Mediated Co-Translational Folding of Recombinant Proteins in Escherichia coli
Abstract
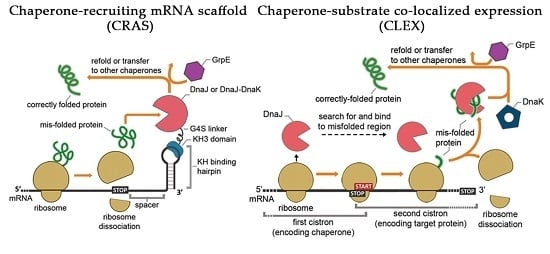
:1. Introduction
2. Results
2.1. The CRAS System Enhances Post-Translational Refolding of Highly Insoluble Recombinant Proteins
2.2. DnaJ Can Function as the Sole Chaperone in CRAS System
2.3. DnaJ-DnaK Chimeric Chaperone Enhances the Efficiency of The Native DnaK System
2.4. In Vivo Monitoring of Chaperone Reactions
2.5. Co-Translational CLEX System Refolding Activity Solubilizes Aggregation-Prone Recombinant Proteins
2.6. Two-Cistron Ordering and DnaJ Represent Primary Factors in Determining CLEX Protein Solubilization Efficiency
2.7. GrpE-Mediated DnaK Release from The Substrate Is Critical for Obtaining Functional Proteins
2.8. The CLEX System Is Not Efficient in Facilitating the Folding of Large Aggregation-Prone Proteins
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Enzymes, and Chemicals
4.2. Construction of Expression Vectors Encoding Chaperones and Target Recombinant Proteins
4.3. Construction of RNA Scaffolds
4.4. Construction of the CLEX System
4.5. Protein Solubilization Test
4.6. Western Blot Analysis
4.7. Protein Solubility Test with in vitro Translated Proteins
4.8. Purification of DnaJ-KH Fusion Proteins
4.9. GFP Complementation Assay
4.10. Electric Mobility Shift Assay
4.11. Activity Assay of Alcohol dehydrogenase 1 (Adh1p)
4.12. Activity Assay of HIV-1 Protease (HIV1-Pr)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Dragosits, M.; Nicklas, D.; Tagkopoulos, I. A synthetic biology approach to self-regulatory recombinant protein production in Escherichia coli. J. Biol. Eng. 2012, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Chou, H.; Ham, T.S.; Lee, T.S.; Keasling, J.D. Metabolic engineering of microorganisms for biofuels production: From bugs to synthetic biology to fuels. Curr. Opin. Biotechnol. 2008, 19, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Baneyx, F.; Mujacic, M. Recombinant protein folding and misfolding in Escherichia coli. Nat. Biotechnol. 2004, 22, 1399–1408. [Google Scholar] [CrossRef] [PubMed]
- Tyedmers, J.; Mogk, A.; Bukau, B. Cellular strategies for controlling protein aggregation. Nat. Rev. Mol. Cell Biol. 2010, 11, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Rajan, R.S.; Illing, M.E.; Bence, N.F.; Kopito, R.R. Specificity in intracellular protein aggregation and inclusion body formation. Proc. Natl. Acad. Sci. USA 2001, 98, 13060–13065. [Google Scholar] [CrossRef] [Green Version]
- Tsumoto, K.; Ejima, D.; Kumagai, I.; Arakawa, T. Practical considerations in refolding proteins from inclusion bodies. Protein Expr. Purif. 2003, 28, 1–8. [Google Scholar] [CrossRef]
- Pryor, K.D.; Leiting, B. High-level expression of soluble protein in Escherichia coli using a His6-tag and maltose-binding-protein double-affinity fusion system. Protein Expr. Purif. 1997, 10, 309–319. [Google Scholar] [CrossRef]
- Marblestone, J.G.; Edavettal, S.C.; Lim, Y.; Lim, P.; Zuo, X.; Butt, T.R. Comparison of SUMO fusion technology with traditional gene fusion systems: Enhanced expression and solubility with SUMO. Protein Sci. 2006, 15, 182–189. [Google Scholar] [CrossRef]
- Brondyk, W.H. Selecting an appropriate method for expressing a recombinant protein. Methods Enzymol. 2009, 463, 131–147. [Google Scholar]
- Kopetzki, E.; Schumacher, G.; Buckel, P. Control of formation of active soluble or inactive insoluble baker’s yeast alpha-glucosidase PI in Escherichia coli by induction and growth conditions. Mol. Gen. Genet. MGG 1989, 216, 149–155. [Google Scholar] [CrossRef]
- Georgiou, G.; Valax, P. Expression of correctly folded proteins in Escherichia coli. Curr. Opin. Biotechnol. 1996, 7, 190–197. [Google Scholar] [CrossRef]
- Sorensen, H.P.; Mortensen, K.K. Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microb. Cell Factories 2005, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- De Marco, A.; Deuerling, E.; Mogk, A.; Tomoyasu, T.; Bukau, B. Chaperone-based procedure to increase yields of soluble recombinant proteins produced in E. coli. BMC Biotechnol. 2007, 7, 32. [Google Scholar] [CrossRef] [PubMed]
- Deuerling, E.; Schulze-Specking, A.; Tomoyasu, T.; Mogk, A.; Bukau, B. Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 1999, 400, 693–696. [Google Scholar] [CrossRef] [PubMed]
- Hartl, F.U.; Hayer-Hartl, M. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science 2002, 295, 1852–1858. [Google Scholar] [CrossRef] [PubMed]
- Feder, M.E.; Hofmann, G.E. Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu. Rev. Physiol. 1999, 61, 243–282. [Google Scholar] [CrossRef] [PubMed]
- Holtkamp, W.; Kokic, G.; Jager, M.; Mittelstaet, J.; Komar, A.A.; Rodnina, M.V. Cotranslational protein folding on the ribosome monitored in real time. Science 2015, 350, 1104–1107. [Google Scholar] [CrossRef] [Green Version]
- Nishihara, K.; Kanemori, M.; Yanagi, H.; Yura, T. Overexpression of trigger factor prevents aggregation of recombinant proteins in Escherichia coli. Appl. Environ. Microb. 2000, 66, 884–889. [Google Scholar] [CrossRef]
- Calloni, G.; Chen, T.; Schermann Sonya, M.; Chang, H.-C.; Genevaux, P.; Agostini, F.; Tartaglia, G.; Hayer-Hartl, M.; Hartl, F.U. DnaK functions as a central hub in the E. coli chaperone network. Cell Rep. 2012, 1, 251–264. [Google Scholar] [CrossRef]
- Mayer, M.P.; Bukau, B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell. Mol. Life Sci. 2005, 62, 670. [Google Scholar] [CrossRef]
- Hendrick, J.P.; Langer, T.; Davis, T.A.; Hartl, F.U.; Wiedmann, M. Control of folding and membrane translocation by binding of the chaperone DnaJ to nascent polypeptides. Proc. Natl. Acad. Sci. USA 1993, 90, 10216–10220. [Google Scholar] [CrossRef] [PubMed]
- Linke, K.; Wolfram, T.; Bussemer, J.; Jakob, U. The roles of the two Zinc binding sites in DnaJ. J. Biol. Chem. 2003, 278, 44457–44466. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.B.; Musunuru, K.; Lewis, H.A.; Burley, S.K.; Darnell, R.B. The tetranucleotide UCAY directs the specific recognition of RNA by the Nova K-homology 3 domain. Proc. Natl. Acad. Sci. USA 2000, 97, 5740–5745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Persic, L.; Horn, I.R.; Rybak, S.; Cattaneo, A.; Hoogenboom, H.R.; Bradbury, A. Single-chain variable fragments selected on the 57-76 p21Ras neutralising epitope from phage antibody libraries recognise the parental protein. FEBS Lett. 1999, 443, 112–116. [Google Scholar] [CrossRef] [Green Version]
- Lim, K.J.; Sung, B.H.; Shin, J.R.; Lee, Y.W.; Kim, D.J.; Yang, K.S.; Kim, S.C. A cancer specific cell-penetrating peptide, BR2, for the efficient delivery of an scFv into cancer cells. PLoS ONE 2013, 8, e66084. [Google Scholar] [CrossRef]
- Vallejo, L.F.; Brokelmann, M.; Marten, S.; Trappe, S.; Cabrera-Crespo, J.; Hoffmann, A.; Gross, G.; Weich, H.A.; Rinas, U. Renaturation and purification of bone morphogenetic protein-2 produced as inclusion bodies in high-cell-density cultures of recombinant Escherichia coli. J. Biotechnol. 2002, 94, 185–194. [Google Scholar] [CrossRef]
- Abella, V.; Scotece, M.; Conde, J.; Pino, J.; Gonzalez-Gay, M.A.; Gómez-Reino, J.J.; Mera, A.; Lago, F.; Gómez, R.; Gualillo, O. Leptin in the interplay of inflammation, metabolism and immune system disorders. Nat. Rev. Rheumatol. 2017, 13, 100. [Google Scholar] [CrossRef] [PubMed]
- Dardeno, T.A.; Chou, S.H.; Moon, H.-S.; Chamberland, J.P.; Fiorenza, C.G.; Mantzoros, C.S. Leptin in human physiology and therapeutics. Front. Neuroendocrinol. 2010, 31, 377–393. [Google Scholar] [CrossRef] [Green Version]
- Volonte, F.; Piubelli, L.; Pollegioni, L. Optimizing HIV-1 protease production in Escherichia coli as fusion protein. Microb. Cell Factories 2011, 10, 53. [Google Scholar] [CrossRef]
- Mainprize, I.L.; Bean, J.D.; Bouwman, C.; Kimber, M.S.; Whitfield, C. The UDP-glucose dehydrogenase of Escherichia coli K-12 displays substrate inhibition by NAD that is relieved by nucleotide triphosphates. J. Biol. Chem. 2013, 288, 23064–23074. [Google Scholar] [CrossRef]
- Sengupta, S.; Jonnalagadda, S.; Goonewardena, L.; Juturu, V. Metabolic engineering of a novel muconic acid biosynthesis pathway via 4-hydroxybenzoic acid in Escherichia coli. Appl. Environ. Microbiol. 2015, 81, 8037–8043. [Google Scholar] [CrossRef] [PubMed]
- Ida, Y.; Furusawa, C.; Hirasawa, T.; Shimizu, H. Stable disruption of ethanol production by deletion of the genes encoding alcohol dehydrogenase isozymes in Saccharomyces cerevisiae. J. Biosci. Bioeng. 2012, 113, 192–195. [Google Scholar] [CrossRef] [PubMed]
- Cho, W.K.; Sohn, U.; Kwak, J.W. Production and in vitro refolding of a single-chain antibody specific for human plasma apolipoprotein A-I. J. Biotechnol. 2000, 77, 169–178. [Google Scholar] [CrossRef]
- Louis, J.M.; Clore, G.M.; Gronenborn, A.M. Autoprocessing of HIV-1 protease is tightly coupled to protein folding. Nat. Struct. Biol. 1999, 6, 868–875. [Google Scholar] [PubMed]
- Siebert, M.; Bechthold, A.; Melzer, M.; May, U.; Berger, U.; Schroder, G.; Schroder, J.; Severin, K.; Heide, L. Ubiquinone biosynthesis. Cloning of the genes coding for chorismate pyruvate-lyase and 4-hydroxybenzoate octaprenyl transferase from Escherichia coli. FEBS Lett. 1992, 307, 347–350. [Google Scholar] [CrossRef]
- Jeong, K.J.; Lee, S.Y. Secretory production of human leptin in Escherichia coli. Biotechnol. Bioeng. 2000, 67, 398–407. [Google Scholar] [CrossRef]
- Wang, L.; Maji, S.K.; Sawaya, M.R.; Eisenberg, D.; Riek, R. Bacterial inclusion bodies contain amyloid-like structure. PLoS Biol. 2008, 6, e195. [Google Scholar] [CrossRef]
- Van Durme, J.; Maurer-Stroh, S.; Gallardo, R.; Wilkinson, H.; Rousseau, F.; Schymkowitz, J. Accurate prediction of DnaK-peptide binding via homology modelling and experimental data. PLoS Comput. Biol. 2009, 5, e1000475. [Google Scholar] [CrossRef]
- Ceroni, A.; Passerini, A.; Vullo, A.; Frasconi, P. DISULFIND: A disulfide bonding state and cysteine connectivity prediction server. Nucleic Acids Res. 2006, 34, W177–W181. [Google Scholar] [CrossRef]
- Srinivasan, S.R.; Gillies, A.T.; Chang, L.; Thompson, A.D.; Gestwicki, J.E. Molecular chaperones DnaK and DnaJ share predicted binding sites on most proteins in the E. coli proteome. Mol. Biosyst. 2012, 8, 2323–2333. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671. [Google Scholar] [CrossRef] [PubMed]
- Blum, P.; Ory, J.; Bauernfeind, J.; Krska, J. Physiological consequences of DnaK and DnaJ overproduction in Escherichia coli. J. Bacteriol. 1992, 174, 7436–7444. [Google Scholar] [CrossRef] [PubMed]
- Nunes, J.M.; Mayer-Hartl, M.; Hartl, F.U.; Müller, D.J. Action of the Hsp70 chaperone system observed with single proteins. Nat. Commun. 2015, 6, 6307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenblum, G.; Cooperman, B.S. Engine out of the chassis: Cell-free protein synthesis and its uses. FEBS Lett. 2014, 588, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Schoner, B.E.; Belagaje, R.M.; Schoner, R.G. Translation of a synthetic two-cistron mRNA in Escherichia coli. Proc. Natl. Acad. Sci. USA 1986, 83, 8506–8510. [Google Scholar] [CrossRef] [PubMed]
- Makoff, A.J.; Smallwood, A.E. The use of two-cistron constructions in improving the expression of a heterologous gene in E. coli. Nucleic Acids Res. 1990, 18, 1711–1718. [Google Scholar] [CrossRef] [PubMed]
- Osterman, I.A.; Evfratov, S.A.; Sergiev, P.V.; Dontsova, O.A. Comparison of mRNA features affecting translation initiation and reinitiation. Nucleic Acids Res. 2013, 41, 474–486. [Google Scholar] [CrossRef] [PubMed]
- Harrison, C.J.; Hayer-Hartl, M.; Liberto, M.D.; Hartl, F.-U.; Kuriyan, J. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science 1997, 276, 431–435. [Google Scholar] [CrossRef]
- Sugimoto, S.; Saruwatari, K.; Higashi, C.; Sonomoto, K. The proper ratio of GrpE to DnaK is important for protein quality control by the DnaK–DnaJ–GrpE chaperone system and for cell division. Microbiology 2008, 154, 1876–1885. [Google Scholar] [CrossRef]
- Perales-Calvo, J.; Giganti, D.; Stirnemann, G.; Garcia-Manyes, S. The force-dependent mechanism of DnaK-mediated mechanical folding. Sci. Adv. 2018, 4, eaaq0243. [Google Scholar] [CrossRef] [Green Version]
- Kyratsous, C.A.; Silverstein, S.J.; DeLong, C.R.; Panagiotidis, C.A. Chaperone-fusion expression plasmid vectors for improved solubility of recombinant proteins in Escherichia coli. Gene 2009, 440, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Alonso, M.; Vera, A.; Villaverde, A. Role of the chaperone DnaK in protein solubility and conformational quality in inclusion body-forming Escherichia coli cells. FEMS Microbiol. Lett. 2007, 273, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Bloom, R.J.; Winkler, S.M.; Smolke, C.D. A quantitative framework for the forward design of synthetic miRNA circuits. Nat. Methods 2014, 11, 1147. [Google Scholar] [CrossRef] [PubMed]
- Lim, F.; Peabody, D.S. RNA recognition site of PP7 coat protein. Nucleic Acids Res. 2002, 30, 4138–4144. [Google Scholar] [CrossRef] [Green Version]
- Ferbitz, L.; Maier, T.; Patzelt, H.; Bukau, B.; Deuerling, E.; Ban, N. Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature 2004, 431, 590. [Google Scholar] [CrossRef]
- Diamant, S.; Ben-Zvi, A.P.; Bukau, B.; Goloubinoff, P. Size-dependent disaggregation of stable protein aggregates by the DnaK chaperone machinery. J. Biol. Chem. 2000, 275, 21107–21113. [Google Scholar] [CrossRef]
- Pierpaoli, E.V.; Sandmeier, E.; Schönfeld, H.-J.; Christen, P. Control of the DnaK chaperone cycle by substoichiometric concentrations of the co-chaperones DnaJ and GrpE. J. Biol. Chem. 1998, 273, 6643–6649. [Google Scholar] [CrossRef]
- Schönfeld, H.-J.; Schmidt, D.; Schröder, H.; Bukau, B. The DnaK chaperone system of Escherichia coli: Quaternary structures and interactions of the DnaK and GrpE components. J. Biol. Chem. 1995, 270, 2183–2189. [Google Scholar] [CrossRef]
- Yu, D.; Ellis, H.M.; Lee, E.-C.; Jenkins, N.A.; Copeland, N.G.; Court, D.L. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 2000, 97, 5978–5983. [Google Scholar] [CrossRef]
- Stewart, E.J.; Åslund, F.; Beckwith, J. Disulfide bond formation in the Escherichia coli cytoplasm: An in vivo role reversal for the thioredoxins. EMBO J. 1998, 17, 5543–5550. [Google Scholar] [CrossRef]
- Klint, J.K.; Senff, S.; Saez, N.J.; Seshadri, R.; Lau, H.Y.; Bende, N.S.; Undheim, E.A.; Rash, L.D.; Mobli, M.; King, G.F. Production of recombinant disulfide-rich venom peptides for structural and functional analysis via expression in the periplasm of E. coli. PLoS ONE 2013, 8, e63865. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.D.; Hatahet, F.; Salo, K.E.; Enlund, E.; Zhang, C.; Ruddock, L.W. Pre-expression of a sulfhydryl oxidase significantly increases the yields of eukaryotic disulfide bond containing proteins expressed in the cytoplasm of E. coli. Microb. Cell Factories 2011, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.T.; Koo, B.-K.; Vu, T.T.T.; Song, J.-A.; Chong, S.-H.; Jeong, B.; Ryu, H.-B.; Moh, S.-H.; Choe, H. Prokaryotic soluble overexpression and purification of bioactive human growth hormone by fusion to thioredoxin, maltose binding protein, and protein disulfide isomerase. PLoS ONE 2014, 9, e89038. [Google Scholar] [CrossRef] [PubMed]
- De Marco, A. Strategies for successful recombinant expression of disulfide bond-dependent proteins in Escherichia coli. Microb. Cell Factories 2009, 8, 26. [Google Scholar] [CrossRef] [PubMed]
- Lobstein, J.; Emrich, C.A.; Jeans, C.; Faulkner, M.; Riggs, P.; Berkmen, M. SHuffle, a novel Escherichia coli protein expression strain capable of correctly folding disulfide bonded proteins in its cytoplasm. Microb. Cell Factories 2012, 11, 753. [Google Scholar] [CrossRef] [PubMed]
- Thomason, L.C.; Costantino, N.; Court, D.L.E. E. coli genome manipulation by P1 transduction. Curr. Protoc. Mol. Biol. 2007. [CrossRef]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef]
- Pichon, C.; du Merle, L.; Lequeutre, I.; Le Bouguenec, C. The AfaR small RNA controls expression of the AfaD-VIII invasin in pathogenic Escherichia coli strains. Nucleic Acids Res. 2013, 41, 5469–5482. [Google Scholar] [CrossRef]
- Salis, H.M.; Mirsky, E.A.; Voigt, C.A. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009, 27, 946–950. [Google Scholar] [CrossRef] [Green Version]
- Teplova, M.; Malinina, L.; Darnell, J.C.; Song, J.; Lu, M.; Abagyan, R.; Musunuru, K.; Teplov, A.; Burley, S.K.; Darnell, R.B.; et al. Protein-RNA and protein-protein recognition by dual KH1/2 domains of the neuronal splicing factor Nova-1. Structure 2011, 19, 930–944. [Google Scholar] [CrossRef]
- Nguyen, H.L.; Nguyen, T.T.; Vu, Q.T.; Le, H.T.; Pham, Y.; Trinh, P.L.; Bui, T.P.; Phan, T.N. An efficient procedure for the expression and purification of HIV-1 protease from inclusion bodies. Protein Expr. Purif. 2015, 116, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.S.; Sung, B.H.; Park, M.K.; Lee, J.H.; Lim, K.J.; Park, S.C.; Kim, S.L.; Kim, H.K.; Sohn, J.H.; Kim, H.M.; et al. Recombinant lipase engineered with amphipathic and coiled-coil peptides. ACS Catal. 2015, 5, 5016–5025. [Google Scholar] [CrossRef]
- Andronescu, M.; Fejes, A.P.; Hutter, F.; Hoos, H.H.; Condon, A. A new algorithm for RNA secondary structure design. J. Mol. Biol. 2004, 336, 607–624. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. On finding all suboptimal foldings of an RNA molecule. Science 1989, 244, 48–52. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.A.; Sung, B.H.; Cho, J.H.; Kim, S.C. Direct expression of antimicrobial peptides in an intact form by a translationally coupled two-cistron expression system. Appl. Environ. Microbiol. 2009, 75, 3980–3986. [Google Scholar] [CrossRef] [PubMed]
- Velappan, N.; Sblattero, D.; Chasteen, L.; Pavlik, P.; Bradbury, A.R.M. Plasmid incompatibility: More compatible than previously thought? Protein Eng. Des. Sel. 2007, 20, 309–313. [Google Scholar] [CrossRef] [PubMed]
- De la Torre, M.; Humanes, M.C.; Olivera, E.R.; Luengo, J.M. Plasmids containing the same origin of replication are useful tools to perform biotechnological studies in Pseudomonas putida U and in E. coli DH10B. Can. J. Biotechnol. 2017, 1, 38–43. [Google Scholar] [CrossRef]
- Cabantous, S.; Terwilliger, T.C.; Waldo, G.S. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol. 2005, 23, 102–107. [Google Scholar] [CrossRef]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bui, L.M.; Geraldi, A.; Nguyen, T.T.; Lee, J.H.; Lee, J.Y.; Cho, B.-K.; Kim, S.C. mRNA Engineering for the Efficient Chaperone-Mediated Co-Translational Folding of Recombinant Proteins in Escherichia coli. Int. J. Mol. Sci. 2019, 20, 3163. https://doi.org/10.3390/ijms20133163
Bui LM, Geraldi A, Nguyen TT, Lee JH, Lee JY, Cho B-K, Kim SC. mRNA Engineering for the Efficient Chaperone-Mediated Co-Translational Folding of Recombinant Proteins in Escherichia coli. International Journal of Molecular Sciences. 2019; 20(13):3163. https://doi.org/10.3390/ijms20133163
Chicago/Turabian StyleBui, Le Minh, Almando Geraldi, Thi Thuy Nguyen, Jun Hyoung Lee, Ju Young Lee, Byung-Kwan Cho, and Sun Chang Kim. 2019. "mRNA Engineering for the Efficient Chaperone-Mediated Co-Translational Folding of Recombinant Proteins in Escherichia coli" International Journal of Molecular Sciences 20, no. 13: 3163. https://doi.org/10.3390/ijms20133163
APA StyleBui, L. M., Geraldi, A., Nguyen, T. T., Lee, J. H., Lee, J. Y., Cho, B. -K., & Kim, S. C. (2019). mRNA Engineering for the Efficient Chaperone-Mediated Co-Translational Folding of Recombinant Proteins in Escherichia coli. International Journal of Molecular Sciences, 20(13), 3163. https://doi.org/10.3390/ijms20133163