The Expression of Thrombospondin-4 Correlates with Disease Severity in Osteoarthritic Knee Cartilage
Abstract
:1. Introduction
2. Results
2.1. Scoring of Articular Cartilage Samples
2.2. TSP-4 Expression and Localization in Human Articular Cartilage
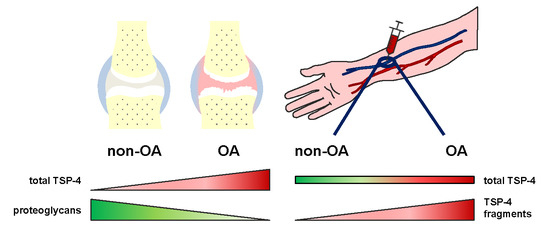
2.3. Total TSP-4 Protein Amount Increases with OA Severity Grade
2.4. Analysis of TSP-4 Anchorage in OA Cartilage
2.5. Gene Expression of Thbs-4 Is Not Increasing During OA
2.6. TSP-4 Levels Were Increased in Serum of OA Patients
3. Discussion
4. Materials and Methods
4.1. Collection of Clinical Species
4.2. Grading of Cartilage Samples
4.3. (Immuno-)Histological Analysis of Cartilage Samples
4.4. Protein Extraction from Articular Cartilage
4.5. SDS-Polyacrylamide Gel Electrophoresis (PAGE) and Immunoblot Analysis
4.6. RNA Isolation and cDNA Synthesis
4.7. Polymerase Chain Reaction and Agrose Gel Electrophoresis
4.8. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ADAMTS | A disintegrin and metalloproteinase with thrombospondin motifs |
| BSA | Bovine serum albumin |
| cDNA | Complementary DNA |
| Col | Collagen |
| COMP | Cartilage oligomeric matrix protein |
| DMEM | Dulbecco’s Modified Eagle’s Medium |
| ECM | Extracellular matrix |
| EDTA | Ethylenediaminetetraacetic acid |
| ELISA | Enzyme-linked immunosorbent assay |
| EtOH | EtOH |
| ER | Endoplasmic reticulum |
| F | Fragment |
| Fig | Figure |
| G | Grade |
| GAPDH | Glycerinaldehyd-3-phosphat-dehydrogenase |
| HC | Healthy controls |
| HRP | Horseradish peroxidase |
| MMP | Matrix metalloproteinase |
| OA | Osteoarthritis |
| P | Pentamer |
| Pat | Patient |
| PBS | Phosphate-buffered saline |
| PCR | Polymerase chain reaction |
| PG | Proteoglycan |
| SDS | Sodium dodecyl sulfate |
| SDS-PAGE | Sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| RT | Room temperature |
| RT-PCR | Reverse transcription polymerase chain reaction |
| TGF-β | Transforming growth factor-β |
| TBS | Tris-buffered saline |
| TSP-4 | Thrombospondin-4 |
| w/v | Weight per volume |
References
- Loeser, R.F.; Goldring, S.R.; Scanzello, C.R.; Goldring, M.B. Osteoarthritis: A disease of the joint as an organ. Arthritis Rheum. 2012, 64, 1697–1707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neogi, T. The epidemiology and impact of pain in osteoarthritis. Osteoarthr. Cartil. 2013, 21, 1145–1153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martel-Pelletier, J.; Boileau, C.; Pelletier, J.-P.; Roughley, P.J. Cartilage in normal and osteoarthritis conditions. Best Pract. Res. Clin. Rheumatol. 2008, 22, 351–384. [Google Scholar] [CrossRef] [PubMed]
- Heijink, A.; Gomoll, A.H.; Madry, H.; Drobnič, M.; Filardo, G.; Espregueira-Mendes, J.; Van Dijk, C.N. Biomechanical considerations in the pathogenesis of osteoarthritis of the knee. Knee Surg. Sports Traumatol. Arthrosc. 2012, 20, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Pratta, M.A.; Yao, W.; Decicco, C.; Tortorella, M.D.; Liu, R.-Q.; Copeland, R.A.; Magolda, R.; Newton, R.C.; Trzaskos, J.M.; Arner, E.C. Aggrecan protects cartilage collagen from proteolytic cleavage. J. Biol. Chem. 2003, 278, 45539–45545. [Google Scholar] [CrossRef] [PubMed]
- Bruckner, P. Suprastructures of extracellular matrices: Paradigms of functions controlled by aggregates rather than molecules. Cell Tissue Res. 2010, 339, 7–18. [Google Scholar] [CrossRef]
- Pullig, O.; Weseloh, G.; Klatt, A.; Wagener, R.; Swoboda, B. Matrilin-3 in human articular cartilage: Increased expression in osteoarthritis. Osteoarthr. Cartil. 2002, 10, 253–263. [Google Scholar] [CrossRef]
- Zack, M.D.; Arner, E.C.; Anglin, C.P.; Alston, J.T.; Malfait, A.M.; Tortorella, M.D. Identification of fibronectin neoepitopes present in human osteoarthritic cartilage. Arthritis Rheum. 2006, 54, 2912–2922. [Google Scholar] [CrossRef] [Green Version]
- Melrose, J.; Fuller, E.S.; Roughley, P.J.; Smith, M.M.; Kerr, B.; Hughes, C.E.; Caterson, B.; Little, C.B. Fragmentation of decorin, biglycan, lumican and keratocan is elevated in degenerate human meniscus, knee and hip articular cartilages compared with age-matched macroscopically normal and control tissues. Arthritis Res. 2008, 10, R79. [Google Scholar] [CrossRef] [Green Version]
- Gebauer, J.M.; Kohler, A.; Dietmar, H.; Gompert, M.; Neundorf, I.; Zaucke, F.; Koch, M.; Baumann, U. COMP and TSP-4 interact specifically with the novel GXKGHR motif only found in fibrillar collagens. Sci. Rep. 2018, 8, 17187. [Google Scholar] [CrossRef]
- Agarwal, P.; Zwolanek, D.; Keene, D.R.; Schulz, J.N.; Blumbach, K.; Heinegard, D.; Zaucke, F.; Paulsson, M.; Krieg, T.; Koch, M.; et al. Collagen XII and XIV, new partners of cartilage oligomeric matrix protein in the skin extracellular matrix suprastructure. J. Biol. Chem. 2012, 287, 22549–22559. [Google Scholar] [CrossRef] [PubMed]
- Mann, H.H.; Ozbek, S.; Engel, J.; Paulsson, M.; Wagener, R. Interactions between the cartilage oligomeric matrix protein and matrilins. Implications for matrix assembly and the pathogenesis of chondrodysplasias. J. Biol. Chem. 2004, 279, 25294–25298. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, K.; Olsson, H.; Mörgelin, M.; Heinegård, D. Cartilage oligomeric matrix protein shows high affinity zinc-dependent interaction with triple helical collagen. J. Biol. Chem. 1998, 273, 20397–20403. [Google Scholar] [CrossRef]
- Di Cesare, P.E.; Chen, F.S.; Moergelin, M.; Carlson, C.S.; Leslie, M.P.; Perris, R.; Fang, C. Matrix–matrix interaction of cartilage oligomeric matrix protein and fibronectin. Matrix Biol. 2002, 21, 461–470. [Google Scholar] [CrossRef]
- Chen, F.H.; Herndon, M.E.; Patel, N.; Hecht, J.T.; Tuan, R.S.; Lawler, J. Interaction of cartilage oligomeric matrix protein/thrombospondin 5 with aggrecan. J. Biol. Chem. 2007, 282, 24591–24598. [Google Scholar] [CrossRef] [PubMed]
- Acharya, C.; Yik, J.H.; Kishore, A.; Van Dinh, V.; Di Cesare, P.E.; Haudenschild, D.R. Cartilage oligomeric matrix protein and its binding partners in the cartilage extracellular matrix: Interaction, regulation and role in chondrogenesis. Matrix Biol. 2014, 37, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.G.; Jordan, J.M.; Vilim, V.; Renner, J.B.; Dragomir, A.D.; Luta, G.; Kraus, V.B. Serum cartilage oligomeric matrix protein reflects osteoarthritis presence and severity: The Johnston County Osteoarthritis Project. Arthritis Rheumatol. 1999, 42, 2356–2364. [Google Scholar] [CrossRef]
- Sakthiswary, R.; Rajalingam, S.; Hussein, H.; Sridharan, R.; Asrul, A.W. Cartilage oligomeric matrix protein (COMP) in rheumatoid arthritis and its correlation with sonographic knee cartilage thickness and disease activity. Clin. Rheumatol. 2017, 36, 2683–2688. [Google Scholar] [CrossRef]
- Liu, F.; Wang, X.; Zhang, X.; Ren, C.; Xin, J. Role of Serum cartilage oligomeric matrix protein (COMP) in the diagnosis of rheumatoid arthritis (RA): A case-control study. J. Int. Med. Res. 2016, 44, 940–949. [Google Scholar] [CrossRef]
- Hesselstrand, R.; Kassner, A.; Heinegard, D.; Saxne, T. COMP: A candidate molecule in the pathogenesis of systemic sclerosis with a potential as a disease marker. Ann. Rheum. Dis. 2008, 67, 1242–1248. [Google Scholar] [CrossRef]
- Norman, G.L.; Gatselis, N.K.; Shums, Z.; Liaskos, C.; Bogdanos, D.P.; Koukoulis, G.K.; Dalekos, G.N. Cartilage oligomeric matrix protein: A novel non-invasive marker for assessing cirrhosis and risk of hepatocellular carcinoma. World J. Hepatol. 2015, 7, 1875–1883. [Google Scholar] [CrossRef] [PubMed]
- Zachou, K.; Gabeta, S.; Shums, Z.; Gatselis, N.K.; Koukoulis, G.K.; Norman, G.L.; Dalekos, G.N. COMP serum levels: A new non-invasive biomarker of liver fibrosis in patients with chronic viral hepatitis. Eur. J. Intern. Med. 2017, 38, 83–88. [Google Scholar] [CrossRef]
- Englund, E.; Bartoschek, M.; Reitsma, B.; Jacobsson, L.; Escudero-Esparza, A.; Orimo, A.; Leandersson, K.; Hagerling, C.; Aspberg, A.; Storm, P.; et al. Cartilage oligomeric matrix protein contributes to the development and metastasis of breast cancer. Oncogene 2016, 35, 5585–5596. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.T.; Liu, X.S.; Zhang, M.; Liu, X.N.; Zhu, F.X.; Zhu, F.M.; Ouyang, S.W.; Li, S.B.; Song, C.L.; Sun, H.M.; et al. Cartilage oligomeric matrix protein is a prognostic factor and biomarker of colon cancer and promotes cell proliferation by activating the Akt pathway. J. Cancer Res. Clin. Oncol. 2018, 144, 1049–1063. [Google Scholar] [CrossRef] [PubMed]
- Ruthard, J.; Hermes, G.; Hartmann, U.; Sengle, G.; Pongratz, G.; Ostendorf, B.; Schneider, M.; Hollriegl, S.; Zaucke, F.; Wagener, R.; et al. Identification of antibodies against extracellular matrix proteins in human osteoarthritis. Biochem. Biophys. Res. Commun. 2018, 503, 1273–1277. [Google Scholar] [CrossRef] [PubMed]
- Sodersten, F.; Ekman, S.; Schmitz, M.; Paulsson, M.; Zaucke, F. Thrombospondin-4 and cartilage oligomeric matrix protein form heterooligomers in equine tendon. Connect. Tissue Res. 2006, 47, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Narouz-Ott, L.; Maurer, P.; Nitsche, D.P.; Smyth, N.; Paulsson, M. Thrombospondin-4 binds specifically to both collagenous and non-collagenous extracellular matrix proteins via its C-terminal domains. J. Biol. Chem. 2000, 275, 37110–37117. [Google Scholar] [CrossRef]
- Jeschke, A.; Bonitz, M.; Simon, M.; Peters, S.; Baum, W.; Schett, G.; Ruether, W.; Niemeier, A.; Schinke, T.; Amling, M. Deficiency of Thrombospondin-4 in Mice Does Not Affect Skeletal Growth or Bone Mass Acquisition, but Causes a Transient Reduction of Articular Cartilage Thickness. PLoS ONE 2015, 10, e0144272. [Google Scholar] [CrossRef]
- Kuttapitiya, A.; Assi, L.; Laing, K.; Hing, C.; Mitchell, P.; Whitley, G.; Harrison, A.; Howe, F.A.; Ejindu, V.; Heron, C.; et al. Microarray analysis of bone marrow lesions in osteoarthritis demonstrates upregulation of genes implicated in osteochondral turnover, neurogenesis and inflammation. Ann. Rheum. Dis. 2017, 76, 1764–1773. [Google Scholar] [CrossRef] [Green Version]
- Tan, F.L.; Moravec, C.S.; Li, J.; Apperson-Hansen, C.; McCarthy, P.M.; Young, J.B.; Bond, M. The gene expression fingerprint of human heart failure. Proc. Natl. Acad. Sci. USA 2002, 99, 11387–11392. [Google Scholar] [CrossRef] [Green Version]
- Frolova, E.G.; Sopko, N.; Blech, L.; Popovic, Z.B.; Li, J.; Vasanji, A.; Drumm, C.; Krukovets, I.; Jain, M.K.; Penn, M.S.; et al. Thrombospondin-4 regulates fibrosis and remodeling of the myocardium in response to pressure overload. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2012, 26, 2363–2373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palao, T.; Medzikovic, L.; Rippe, C.; Wanga, S.; Al-Mardini, C.; van Weert, A.; de Vos, J.; van der Wel, N.N.; van Veen, H.A.; van Bavel, E.T.; et al. Thrombospondin-4 mediates cardiovascular remodelling in angiotensin II-induced hypertension. Cardiovasc. Pathol. Off. J. Soc. Cardiovasc. Pathol. 2018, 35, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Dunkle, E.T.; Zaucke, F.; Clegg, D.O. Thrombospondin-4 and matrix three-dimensionality in axon outgrowth and adhesion in the developing retina. Exp. Eye Res. 2007, 84, 707–717. [Google Scholar] [CrossRef]
- Aigner, T.; Zhu, Y.; Chansky, H.H.; Matsen, F.A., 3rd; Maloney, W.J.; Sandell, L.J. Reexpression of type IIA procollagen by adult articular chondrocytes in osteoarthritic cartilage. Arthritis Rheum. 1999, 42, 1443–1450. [Google Scholar] [CrossRef] [Green Version]
- Koelling, S.; Clauditz, T.S.; Kaste, M.; Miosge, N. Cartilage oligomeric matrix protein is involved in human limb development and in the pathogenesis of osteoarthritis. Arthritis Res. 2006, 8, R56. [Google Scholar] [CrossRef] [PubMed]
- Kempson, G.E. Relationship between the tensile properties of articular cartilage from the human knee and age. Ann. Rheum. Dis. 1982, 41, 508–511. [Google Scholar] [CrossRef]
- Kempson, G.E.; Freeman, M.A.; Swanson, S.A. Tensile properties of articular cartilage. Nature 1968, 220, 1127–1128. [Google Scholar] [CrossRef] [PubMed]
- Sophia Fox, A.J.; Bedi, A.; Rodeo, S.A. The basic science of articular cartilage: Structure, composition, and function. Sports Health 2009, 1, 461–468. [Google Scholar] [CrossRef]
- Giannoni, P.; Siegrist, M.; Hunziker, E.B.; Wong, M. The mechanosensitivity of cartilage oligomeric matrix protein (COMP). Biorheology 2003, 40, 101–109. [Google Scholar] [PubMed]
- Cingolani, O.H.; Kirk, J.A.; Seo, K.; Koitabashi, N.; Lee, D.I.; Ramirez-Correa, G.; Bedja, D.; Barth, A.S.; Moens, A.L.; Kass, D.A. Thrombospondin-4 is required for stretch-mediated contractility augmentation in cardiac muscle. Circ. Res. 2011, 109, 1410–1414. [Google Scholar] [CrossRef]
- Islam, A.; Mbimba, T.; Younesi, M.; Akkus, O. Effects of substrate stiffness on the tenoinduction of human mesenchymal stem cells. Acta Biomater. 2017, 58, 244–253. [Google Scholar] [CrossRef]
- Mustonen, E.; Aro, J.; Puhakka, J.; Ilves, M.; Soini, Y.; Leskinen, H.; Ruskoaho, H.; Rysa, J. Thrombospondin-4 expression is rapidly upregulated by cardiac overload. Biochem. Biophys. Res. Commun. 2008, 373, 186–191. [Google Scholar] [CrossRef]
- Subramanian, A.; Schilling, T.F. Thrombospondin-4 controls matrix assembly during development and repair of myotendinous junctions. eLife 2014, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuznetsova, S.A.; Issa, P.; Perruccio, E.M.; Zeng, B.; Sipes, J.M.; Ward, Y.; Seyfried, N.T.; Fielder, H.L.; Day, A.J.; Wight, T.N.; et al. Versican-thrombospondin-1 binding in vitro and colocalization in microfibrils induced by inflammation on vascular smooth muscle cells. J. Cell Sci. 2006, 119 Pt 21, 4499–4509. [Google Scholar] [CrossRef] [Green Version]
- Schminke, B.; Frese, J.; Bode, C.; Goldring, M.B.; Miosge, N. Laminins and Nidogens in the Pericellular Matrix of Chondrocytes: Their Role in Osteoarthritis and Chondrogenic Differentiation. Am. J. Pathol. 2016, 186, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Kruegel, J.; Sadowski, B.; Miosge, N. Nidogen-1 and nidogen-2 in healthy human cartilage and in late-stage osteoarthritis cartilage. Arthritis Rheum. 2008, 58, 1422–1432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guilak, F.; Nims, R.; Dicks, A.; Wu, C.-L.; Meulenbelt, I. Osteoarthritis as a disease of the cartilage pericellular matrix. Matrix Biol. 2018, 71–72, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Ronziere, M.C.; Ricard-Blum, S.; Tiollier, J.; Hartmann, D.J.; Garrone, R.; Herbage, D. Comparative analysis of collagens solubilized from human foetal, and normal and osteoarthritic adult articular cartilage, with emphasis on type VI collagen. Biochim. Biophys. Acta 1990, 1038, 222–230. [Google Scholar] [CrossRef]
- Nugent, A.E.; Speicher, D.M.; Gradisar, I.; McBurney, D.L.; Baraga, A.; Doane, K.J.; Horton, W.E., Jr. Advanced osteoarthritis in humans is associated with altered collagen VI expression and upregulation of ER-stress markers Grp78 and bag-1. J. Histochem. Cytochem. Off. J. Histochem. Soc. 2009, 57, 923–931. [Google Scholar] [CrossRef] [PubMed]
- Pullig, O.; Weseloh, G.; Swoboda, B. Expression of type VI collagen in normal and osteoarthritic human cartilage. Osteoarthr. Cartil. 1999, 7, 191–202. [Google Scholar] [CrossRef] [Green Version]
- Chang, J.; Poole, C.A. Sequestration of type VI collagen in the pericellular microenvironment of adult chrondrocytes cultured in agarose. Osteoarthr. Cartil. 1996, 4, 275–285. [Google Scholar] [CrossRef]
- Brachvogel, B.; Zaucke, F.; Dave, K.; Norris, E.L.; Stermann, J.; Dayakli, M.; Koch, M.; Gorman, J.J.; Bateman, J.F.; Wilson, R. Comparative proteomic analysis of normal and collagen IX null mouse cartilage reveals altered extracellular matrix composition and novel components of the collagen IX interactome. J. Biol. Chem. 2013, 288, 13481–13492. [Google Scholar] [CrossRef]
- Budde, B.; Blumbach, K.; Ylöstalo, J.; Zaucke, F.; Ehlen, H.W.; Wagener, R.; Ala-Kokko, L.; Paulsson, M.; Bruckner, P.; Grässel, S. Altered integration of matrilin-3 into cartilage extracellular matrix in the absence of collagen IX. Mol. Cell. Biol. 2005, 25, 10465–10478. [Google Scholar] [CrossRef] [PubMed]
- Groma, G.; Xin, W.; Grskovic, I.; Niehoff, A.; Brachvogel, B.; Paulsson, M.; Zaucke, F. Abnormal bone quality in cartilage oligomeric matrix protein and matrilin 3 double-deficient mice caused by increased tissue inhibitor of metalloproteinases 3 deposition and delayed aggrecan degradation. Arthritis Rheum. 2012, 64, 2644–2654. [Google Scholar] [CrossRef] [Green Version]
- Lynch, J.M.; Maillet, M.; Vanhoutte, D.; Schloemer, A.; Sargent, M.A.; Blair, N.S.; Lynch, K.A.; Okada, T.; Aronow, B.J.; Osinska, H.; et al. A thrombospondin-dependent pathway for a protective ER stress response. Cell 2012, 149, 1257–1268. [Google Scholar] [CrossRef] [PubMed]
- Brody, M.J.; Vanhoutte, D.; Schips, T.G.; Boyer, J.G.; Bakshi, C.V.; Sargent, M.A.; York, A.J.; Molkentin, J.D. Defective Flux of Thrombospondin-4 through the Secretory Pathway Impairs Cardiomyocyte Membrane Stability and Causes Cardiomyopathy. Mol. Cell. Biol. 2018. [Google Scholar] [CrossRef]
- Rosini, S.; Pugh, N.; Bonna, A.M.; Hulmes, D.J.S.; Farndale, R.W.; Adams, J.C. Thrombospondin-1 promotes matrix homeostasis by interacting with collagen and lysyl oxidase precursors and collagen cross-linking sites. Sci. Signal. 2018, 11, 532. [Google Scholar] [CrossRef] [PubMed]
- Schulz, J.N.; Nuchel, J.; Niehoff, A.; Bloch, W.; Schonborn, K.; Hayashi, S.; Kamper, M.; Brinckmann, J.; Plomann, M.; Paulsson, M.; et al. COMP-assisted collagen secretion--a novel intracellular function required for fibrosis. J. Cell Sci. 2016, 129, 706–716. [Google Scholar] [CrossRef]
- von der Mark, K.; Frischholz, S.; Aigner, T.; Beier, F.; Belke, J.; Erdmann, S.; Burkhardt, H. Upregulation of type X collagen expression in osteoarthritic cartilage. Acta Orthop. Scand. Suppl. 1995, 266, 125–129. [Google Scholar] [CrossRef]
- Lorenzo, P.; Aspberg, A.; Saxne, T.; Onnerfjord, P. Quantification of cartilage oligomeric matrix protein (COMP) and a COMP neoepitope in synovial fluid of patients with different joint disorders by novel automated assays. Osteoarthr. Cartil. 2017, 25, 1436–1442. [Google Scholar] [CrossRef]
- Skioldebrand, E.; Ekman, S.; Mattsson Hulten, L.; Svala, E.; Bjorkman, K.; Lindahl, A.; Lundqvist, A.; Onnerfjord, P.; Sihlbom, C.; Ruetschi, U. Cartilage oligomeric matrix protein neoepitope in the synovial fluid of horses with acute lameness: A new biomarker for the early stages of osteoarthritis. Equine Vet. J. 2017, 49, 662–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, L.; Guo, D.; Homandberg, G.A. Fibronectin fragments mediate matrix metalloproteinase upregulation and cartilage damage through proline rich tyrosine kinase 2, c-src, NF-kappaB and protein kinase Cdelta. Osteoarthr. Cartil. 2009, 17, 1385–1392. [Google Scholar] [CrossRef]
- Klatt, A.R.; Klinger, G.; Paul-Klausch, B.; Kühn, G.; Renno, J.H.; Wagener, R.; Paulsson, M.; Schmidt, J.; Malchau, G.; Wielckens, K. Matrilin-3 activates the expression of osteoarthritis-associated genes in primary human chondrocytes. FEBS Lett. 2009, 583, 3611–3617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, G.Q.; Chen, A.B.; Li, W.; Song, J.H.; Gao, C.Y. High MMP-1, MMP-2, and MMP-9 protein levels in osteoarthritis. Genet. Mol. Res. GMR 2015, 14, 14811–14822. [Google Scholar] [CrossRef]
- Liu, C.J. The role of ADAMTS-7 and ADAMTS-12 in the pathogenesis of arthritis. Nat. Clin. Pract. Rheumatol. 2009, 5, 38–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blaney Davidson, E.N.; Remst, D.F.; Vitters, E.L.; van Beuningen, H.M.; Blom, A.B.; Goumans, M.J.; van den Berg, W.B.; van der Kraan, P.M. Increase in ALK1/ALK5 ratio as a cause for elevated MMP-13 expression in osteoarthritis in humans and mice. J. Immunol. 2009, 182, 7937–7945. [Google Scholar] [CrossRef] [PubMed]
- Ganu, V.; Goldberg, R.; Peppard, J.; Rediske, J.; Melton, R.; Hu, S.I.; Wang, W.; Duvander, C.; Heinegard, D. Inhibition of interleukin-1alpha-induced cartilage oligomeric matrix protein degradation in bovine articular cartilage by matrix metalloproteinase inhibitors: Potential role for matrix metalloproteinases in the generation of cartilage oligomeric matrix protein fragments in arthritic synovial fluid. Arthritis Rheum. 1998, 41, 2143–2151. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-j.; Kong, W.; Ilalov, K.; Yu, S.; Xu, K.; Prazak, L.; Fajardo, M.; Sehgal, B.; Di Cesare, P.E. ADAMTS-7: A metalloproteinase that directly binds to and degrades cartilage oligomeric matrix protein. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2006, 20, 988–990. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-j.; Kong, W.; Xu, K.; Luan, Y.; Ilalov, K.; Sehgal, B.; Yu, S.; Howell, R.D.; Di Cesare, P.E. ADAMTS-12 associates with and degrades cartilage oligomeric matrix protein. J. Biol. Chem. 2006, 281, 15800–15808. [Google Scholar] [CrossRef]
- Büchele, G.; Günther, K.P.; Brenner, H.; Puhl, W.; Stürmer, T.; Rothenbacher, D.; Brenner, R.E. Osteoarthritis-patterns, cardio-metabolic risk factors and risk of all-cause mortality: 20 years follow-up in patients after hip or knee replacement. Sci. Rep. 2018, 8, 5253. [Google Scholar] [CrossRef] [PubMed]
- Pritzker, K.P.; Gay, S.; Jimenez, S.A.; Ostergaard, K.; Pelletier, J.P.; Revell, P.A.; Salter, D.; van den Berg, W.B. Osteoarthritis cartilage histopathology: Grading and staging. Osteoarthr. Cartil. 2006, 14, 13–29. [Google Scholar] [CrossRef] [PubMed]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maly, K.; Schaible, I.; Riegger, J.; Brenner, R.E.; Meurer, A.; Zaucke, F. The Expression of Thrombospondin-4 Correlates with Disease Severity in Osteoarthritic Knee Cartilage. Int. J. Mol. Sci. 2019, 20, 447. https://doi.org/10.3390/ijms20020447
Maly K, Schaible I, Riegger J, Brenner RE, Meurer A, Zaucke F. The Expression of Thrombospondin-4 Correlates with Disease Severity in Osteoarthritic Knee Cartilage. International Journal of Molecular Sciences. 2019; 20(2):447. https://doi.org/10.3390/ijms20020447
Chicago/Turabian StyleMaly, Kathrin, Inna Schaible, Jana Riegger, Rolf E. Brenner, Andrea Meurer, and Frank Zaucke. 2019. "The Expression of Thrombospondin-4 Correlates with Disease Severity in Osteoarthritic Knee Cartilage" International Journal of Molecular Sciences 20, no. 2: 447. https://doi.org/10.3390/ijms20020447
APA StyleMaly, K., Schaible, I., Riegger, J., Brenner, R. E., Meurer, A., & Zaucke, F. (2019). The Expression of Thrombospondin-4 Correlates with Disease Severity in Osteoarthritic Knee Cartilage. International Journal of Molecular Sciences, 20(2), 447. https://doi.org/10.3390/ijms20020447