Effects of Dietary Lipid Composition and Fatty Acid Desaturase 2 Expression in Broodstock Gilthead Sea Bream on Lipid Metabolism-Related Genes and Methylation of the fads2 Gene Promoter in Their Offspring
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Fish Feeding
4.2. Molecular Studies
4.2.1. Quantification of fads2 Expression of Brood Fish
4.2.2. Digital Droplet PCR Analysis for Absolute Gene Expression in Juveniles
4.2.3. DNA Methylation Analysis
4.3. Statistical Analysis
5. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization. The State of World Fisheries and Aquaculture 2018—Meeting the Sustainable Development Goals; Food and Agriculture Organization: Rome, Italy, 2018. [Google Scholar]
- Council, N.R. Critical Role of Animal Science Research in Food Security and Sustainability; National Academies Press: Washington, DC, USA, 2015. [Google Scholar]
- Montero, D.; Izquierdo, M. Welfare and health of fish fed vegetable oils as alternative lipid sources to fish oil. In Fish Oil Replacement and Alternative Lipid Sources in Aquaculture Feeds; Turchini, G.M., Ng, W.K., Tocher, D.R., Eds.; CRC Press: Boca Raton, FL, USA, 2010; pp. 439–485. [Google Scholar]
- Torrecillas, S.; Román, L.; Rivero-Ramírez, F.; Caballero, M.J.; Pascual, C.; Robaina, L.; Izquierdo, M.S.; Acosta, F.; Montero, D. Supplementation of arachidonic acid rich oil in European sea bass juveniles (Dicentrarchus labrax) diets: Effects on leucocytes and plasma fatty acid profiles, selected immune parameters and circulating prostaglandins levels. Fish Shellfish Immunol. 2017, 64, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Torrecillas, S.; Mompel, D.; Caballero, M.J.; Montero, D.; Merrifield, D.; Rodiles, A.; Robaina, L.; Zamorano, M.J.; Karalazos, V.; Kaushik, S.; et al. Effect of fishmeal and fish oil replacement by vegetable meals and oils on gut health of European sea bass (Dicentrarchus labrax). Aquaculture 2017, 468, 386–398. [Google Scholar] [CrossRef]
- Dawood, M.A.O.; Koshio, S.; Esteban, M.Á. Beneficial roles of feed additives as immunostimulants in aquaculture: A review. Rev. Aquac. 2018, 10, 950–974. [Google Scholar] [CrossRef]
- Hardy, R.W. Utilization of plant proteins in fish diets: Effects of global demand and supplies of fishmeal. Aquac. Res. 2010, 41, 770–776. [Google Scholar] [CrossRef]
- Turchini, G.M.; Torstensen, B.E.; Ng, W.K. Fish oil replacement in finfish nutrition. Rev. Aquac. 2009, 1, 10–57. [Google Scholar] [CrossRef]
- Eroldoğan, T.O.; Yılmaz, A.H.; Turchini, G.M.; Arslan, M.; Sirkecioğlu, N.A.; Engin, K.; Özşahinoğlu, I.; Mumoğullarında, P. Fatty acid metabolism in European sea bass (Dicentrarchus labrax): Effects of n-6 PUFA and MUFA in fish oil replaced diets. Fish Physiol. Biochem. 2013, 39, 941–955. [Google Scholar] [CrossRef]
- Rosenlund, G.; Corraze, G.; Izquierdo, M.; Torstensen, B.E. The effects of fish oil replacement on nutritional and organoleptic qualities of farmed fish. In Fish Oil Replacement and Alternative Lipid Sources in Aquaculture Feeds; Turchini, G.M., Ng, W.K., Tocher, D.R., Eds.; CRC Press: Boca Raton, FL, USA, 2010; pp. 487–522. [Google Scholar] [CrossRef]
- Izquierdo, M.S.; Montero, D.; Robaina, L.; Caballero, M.J.; Rosenlund, G.; Ginés, R. Alterations in fillet fatty acid profile and flesh quality in gilthead seabream (Sparus aurata) fed vegetable oils for a long term period. Recovery of fatty acid profiles by fish oil feeding. Aquaculture 2005, 250, 431–444. [Google Scholar] [CrossRef] [Green Version]
- Yılmaz, H.A.; Corraze, G.; Panserat, S.; Eroldoğan, O.T. Effects of alternate feeding with different lipid sources on fatty acid composition and bioconversion in European sea bass (Dicentrarchus labrax). Aquaculture 2016, 464, 28–36. [Google Scholar] [CrossRef]
- Jobling, M. National Research Council (NRC): Nutrient requirements of fish and shrimp. Aquac. Int. 2012, 20, 601–602. [Google Scholar] [CrossRef]
- Vagner, M.; Santigosa, E. Characterization and modulation of gene expression and enzymatic activity of delta-6 desaturase in teleosts: A review. Aquaculture 2011, 315, 131–143. [Google Scholar] [CrossRef]
- Izquierdo, M.S.; Robaina, L.; Juárez-Carrillo, E.; Oliva, V.; Hernández-Cruz, C.; Afonso, J. Regulation of growth, fatty acid composition and delta 6 desaturase expression by dietary lipids in gilthead seabream larvae (Sparus aurata). Fish Physiol. Biochem. 2008, 34, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Monroig, Ó.; Navarro, J.C.; Tocher, D.R. Long-chain polyunsaturated fatty acids in fish: Recent advances on desaturases and elongases involved in their biosyntesis. Av. Nutr. Acuícola 2011, 11, 257–283. [Google Scholar]
- Matsuzaka, T.; Shimano, H. Elovl6: A new player in fatty acid metabolism and insulin sensitivity. J. Mol. Med. 2009, 87, 379–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, S.D. Polyunsaturated Fatty Acid Regulation of Gene Transcription: A Molecular Mechanism to Improve the Metabolic Syndrome. J. Nutr. 2001, 131, 1129–1132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bell, G.; Koppe, W. Lipids in aquafeeds. In Fish Oil Replacement and Alternative Lipid Sources in Aquaculture Feeds; Turchini, G.M., Ng, W.K., Tocher, D., Eds.; CRC Press: Boca Raton, FL, USA, 2010; pp. 21–60. [Google Scholar]
- Raclot, T.; Groscolas, R.; Langin, D.; Ferre, P. Site-specific regulation of gene expression by n-3 polyunsaturated fatty acids in rat white adipose tissues. J. Lipid Res. 1997, 38, 1963–1972. [Google Scholar] [PubMed]
- Sargent, J.; Tocher, D.; Bell, J. The lipids. In Fish Nutrition; Halver, J.E., Hardy, R.W., Eds.; Academic Press: San Diego, CA, USA, 2002; pp. 181–257. [Google Scholar]
- Kjaer, M.A.; Todorcevic, M.; Torstensen, B.E.; Vegusdal, A.; Ruyter, B. Dietary n-3 HUFA affects mitochondrial fatty acid beta-oxidation capacity and susceptibility to oxidative stress in Atlantic salmon. Lipids 2008, 43, 813–827. [Google Scholar] [CrossRef]
- Leaver, M.J.; Villeneuve, L.A.; Obach, A.; Jensen, L.; Bron, J.E.; Tocher, D.R.; Taggart, J.B. Functional genomics reveals increases in cholesterol biosynthetic genes and highly unsaturated fatty acid biosynthesis after dietary substitution of fish oil with vegetable oils in Atlantic salmon (Salmo salar). BMC Genom. 2008, 9, 299. [Google Scholar] [CrossRef] [Green Version]
- Vestergren, A.S.; Trattner, S.; Pan, J.; Johnsson, P.; Kamal-Eldin, A.; Brännäs, E.; Moazzami, A.A.; Pickova, J. The effect of combining linseed oil and sesamin on the fatty acid composition in white muscle and on expression of lipid-related genes in white muscle and liver of rainbow trout (Oncorhynchus mykiss). Aquac. Int. 2012, 21, 843–859. [Google Scholar] [CrossRef]
- Xue, X.; Hixson, S.M.; Hori, T.S.; Booman, M.; Parrish, C.C.; Anderson, D.M.; Rise, M.L. Atlantic salmon (Salmo salar) liver transcriptome response to diets containing Camelina sativa products. Comp. Biochem. Physiol. D Genom. Proteom. 2015, 14, 1–15. [Google Scholar] [CrossRef]
- Leaver, M.J.; Bautista, J.M.; Bjornsson, B.T.; Jonsson, E.; Krey, G.; Tocher, D.R.; Torstensen, B.E. Towards fish lipid nutrigenomics: Current state and prospects for fin-fish aquaculture. Rev. Fish. Sci. 2008, 16, 73–94. [Google Scholar] [CrossRef] [Green Version]
- Corbin, K.D. Metabolic Syndrome, Obesity, and Diabetes. In Nutrition in Epigenetics; Niculescu, M.D., Haggarty, P., Eds.; Wiley-Blackwell: Oxford, UK, 2011; pp. 145–164. [Google Scholar] [CrossRef]
- Lillycrop, K.A.; Phillips, E.S.; Torrens, C.; Hanson, M.A.; Jackson, A.A.; Burdge, G.C. Feeding pregnant rats a protein-restricted diet persistently alters the methylation of specific cytosines in the hepatic PPARα promoter of the offspring. Br. J. Nutr. 2008, 100, 278–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishikawa, T.O.; Herschman, H.R. Two inducible, functional cyclooxygenase-2 genes are present in the rainbow trout genome. J. Cell. Biochem. 2007, 102, 1486–1492. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo, M.S.; Turkmen, S.; Montero, D.; Zamorano, M.J.; Afonso, J.M.; Karalazos, V.; Fernández-Palacios, H. Nutritional programming through broodstock diets to improve utilization of very low fishmeal and fish oil diets in gilthead sea bream. Aquaculture 2015, 449, 18–26. [Google Scholar] [CrossRef]
- Turkmen, S.; Zamorano, M.J.; Fernández-Palacios, H.; Hernández-Cruz, C.M.; Montero, D.; Robaina, L.; Izquierdo, M. Parental nutritional programming and a reminder during juvenile stage affect growth, lipid metabolism and utilisation in later developmental stages of a marine teleost, the gilthead sea bream (Sparus aurata). Br. J. Nutr. 2017, 118, 500–512. [Google Scholar] [CrossRef] [Green Version]
- Burdge, G.C.; Lillycrop, K.A. Nutrition, epigenetics, and developmental plasticity: Implications for understanding human disease. Annu. Rev. Nutr. 2010, 30, 315–339. [Google Scholar] [CrossRef]
- Lillycrop, K.A.; Burdge, G.C. Chapter 13—Interactions Between Polyunsaturated Fatty Acids and the Epigenome. In Polyunsaturated Fatty Acid Metabolism; AOCS Press: Urbana, IL, USA, 2018; pp. 225–239. [Google Scholar] [CrossRef]
- Hoile, S.P.; Irvine, N.A.; Kelsall, C.J.; Sibbons, C.; Feunteun, A.; Collister, A.; Torrens, C.; Calder, P.C.; Hanson, M.A.; Lillycrop, K.A.; et al. Maternal fat intake in rats alters 20:4n-6 and 22:6n-3 status and the epigenetic regulation of Fads2 in offspring liver. J. Nutr. Biochem. 2013, 24, 1213–1220. [Google Scholar] [CrossRef] [Green Version]
- Panserat, S.; Marandel, L.; Seiliez, I.; Skiba-Cassy, S. New Insights on Intermediary Metabolism for a Better Understanding of Nutrition in Teleosts. Annu. Rev. Anim. Biosci. 2019, 7. [Google Scholar] [CrossRef]
- Xu, H.; Dong, X.; Ai, Q.; Mai, K.; Xu, W.; Zhang, Y.; Zuo, R. Regulation of Tissue LC-PUFA Contents, Δ6 Fatty Acyl Desaturase (FADS2) Gene Expression and the Methylation of the Putative FADS2 Gene Promoter by Different Dietary Fatty Acid Profiles in Japanese Seabass (Lateolabrax japonicus). PLoS ONE 2014, 9, e87726. [Google Scholar] [CrossRef]
- Geay, F.; Zambonino-Infante, J.; Reinhardt, R.; Kuhl, H.; Santigosa, E.; Cahu, C.; Mazurais, D. Characteristics of fads2 gene expression and putative promoter in European sea bass (Dicentrarchus labrax): Comparison with salmonid species and analysis of CpG methylation. Mar. Genom. 2012, 5, 7–13. [Google Scholar] [CrossRef] [Green Version]
- Pauletto, M.; Manousaki, T.; Ferraresso, S.; Babbucci, M.; Tsakogiannis, A.; Louro, B.; Vitulo, N.; Quoc, V.H.; Carraro, R.; Bertotto, D. Genomic analysis of Sparus aurata reveals the evolutionary dynamics of sex-biased genes in a sequential hermaphrodite fish. Commun. Biol. 2018, 1, 119. [Google Scholar] [CrossRef]
- Torrecillas, S.; Robaina, L.; Caballero, M.J.; Montero, D.; Calandra, G.; Mompel, D.; Karalazos, V.; Kaushik, S.; Izquierdo, M.S. Combined replacement of fishmeal and fish oil in European sea bass (Dicentrarchus labrax): Production performance, tissue composition and liver morphology. Aquaculture 2017, 474, 101–112. [Google Scholar] [CrossRef]
- Izquierdo, M.S.; Fernandez-Palacios, H.; Tacon, A.G.J. Effect of broodstock nutrition on reproductive performance of fish. Aquaculture 2001, 197, 25–42. [Google Scholar] [CrossRef]
- Burdge, G.C.; Calder, P.C. Dietary α-linolenic acid and health-related outcomes: A metabolic perspective. Nutr. Res. Rev. 2006, 19, 26–52. [Google Scholar] [CrossRef] [PubMed]
- Childs, C.E.; Hoile, S.P.; Burdge, G.C.; Calder, P.C. Changes in rat n-3 and n-6 fatty acid composition during pregnancy are associated with progesterone concentrations and hepatic FADS2 expression. Prostaglandins Leukot. Essent. Fat. Acids 2012, 86, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Lupu, D.S.; Cheatham, C.L.; Corbin, K.D.; Niculescu, M.D. Genetic and epigenetic transgenerational implications related to omega-3 fatty acids. Part I: Maternal FADS2 genotype and DNA methylation correlate with polyunsaturated fatty acid status in toddlers: An exploratory analysis. Nutr. Res. 2015, 35, 939–947. [Google Scholar] [CrossRef]
- Moon, Y.-A.; Ochoa, C.R.; Mitsche, M.A.; Hammer, R.E.; Horton, J.D. Deletion of ELOVL6 blocks the synthesis of oleic acid but does not prevent the development of fatty liver or insulin resistance. J. Lipid Res. 2014, 55, 2597–2605. [Google Scholar] [CrossRef] [Green Version]
- Matsuzaka, T.; Shimano, H.; Yahagi, N.; Kato, T.; Atsumi, A.; Yamamoto, T.; Inoue, N.; Ishikawa, M.; Okada, S.; Ishigaki, N.; et al. Crucial role of a long-chain fatty acid elongase, Elovl6, in obesity-induced insulin resistance. Nat. Med. 2007, 13, 1193. [Google Scholar] [CrossRef]
- Turkmen, S.; Hernandez-Cruz, C.M.; Zamorano, M.J.; Fernandez-Palacios, H.; Montero, D.; Afonso, J.M.; Izquierdo, M. Long-chain PUFA profiles in parental diets induce long-term effects on growth, fatty acid profiles, expression of fatty acid desaturase 2 and selected immune system-related genes in the offspring of gilthead seabream. Br. J. Nutr. 2019, 1–14. [Google Scholar] [CrossRef]
- Thomassen, M.S.; Rein, D.; Berge, G.M.; Østbye, T.-K.; Ruyter, B. High dietary EPA does not inhibit ∆5 and ∆6 desaturases in Atlantic salmon (Salmo salar L.) fed rapeseed oil diets. Aquaculture 2012, 360, 78–85. [Google Scholar] [CrossRef]
- Gibson, R.A.; Neumann, M.A.; Lien, E.L.; Boyd, K.A.; Tu, W.C. Docosahexaenoic acid synthesis from alpha-linolenic acid is inhibited by diets high in polyunsaturated fatty acids. Prostaglandins Leukot. Essent. Fat. Acids 2013, 88, 139–146. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Sokal, R.R.; Rohlf, F.J. The Principles and Practice of Statistics in Biological Research; WH Freeman and Company: San Francisco, CA, USA, 1969. [Google Scholar]
- Benedito-Palos, L.; Ballester-Lozano, G.; Pérez-Sánchez, J. Wide-gene expression analysis of lipid-relevant genes in nutritionally challenged gilthead sea bream (Sparus aurata). Gene 2014, 547, 34–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sepulcre, M.P.; López-Castejón, G.; Meseguer, J.; Mulero, V. The activation of gilthead seabream professional phagocytes by different PAMPs underlines the behavioural diversity of the main innate immune cells of bony fish. Mol. Immunol. 2007, 44, 2009–2016. [Google Scholar] [CrossRef]
- Boukouvala, E.; Leaver, M.J.; Favre-Krey, L.; Theodoridou, M.; Krey, G. Molecular characterization of a gilthead sea bream (Sparus aurata) muscle tissue cDNA for carnitine palmitoyltransferase 1B (CPT1B). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2010, 157, 189–197. [Google Scholar] [CrossRef] [Green Version]
- Santos, C.R.A.; Power, D.M.; Kille, P.; Llewellyn, L.; Ramsurn, V.; Wigham, T.; Sweeney, G.E. Cloning and Sequencing of a Full-Length Sea Bream (Sparus aurata) β-Actin cDNA. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 1997, 117, 185–189. [Google Scholar] [CrossRef]



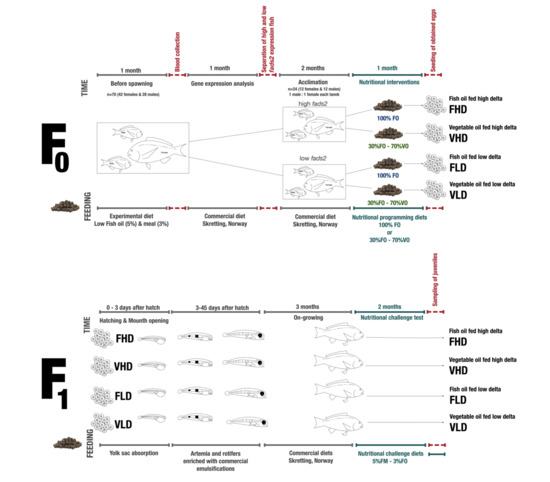
 , and experimental periods when fish were fed with different diets are shown as
, and experimental periods when fish were fed with different diets are shown as  .
.
 , and experimental periods when fish were fed with different diets are shown as
, and experimental periods when fish were fed with different diets are shown as  .
.
| Main Ingredients (%) | % | Proximate Composition | (% Dry Matter) |
|---|---|---|---|
| Fish meal SA 1 68 super prime 1 | 5.00 | Crude lipids | 21.7 |
| Fish meal alternative protein sources 2 | 54.50 | Crude protein | 45.1 |
| Rapeseed meal cake | 11.30 | Moisture | 9.0 |
| Wheat | 6.89 | Ash | 5.4 |
| Fish oil SA 1 | 3.00 | ||
| Vegetable oil mix 3 | 13.00 | Gross Energy (MJ/kg, as is) | 22.5 |
| % Total Fatty Acids | % Total Fatty Acids | ||
| 14:0 | 6.6 | 18:3n-3 | 11.8 |
| 14:1n-5 | 0.1 | 18:4n-3 | 0.4 |
| 15:0 | 0.1 | 18:4n-1 | 0.0 |
| 16:0ISO | 0.0 | 20:0 | 0.4 |
| 16:0 | 12.3 | 20:1n-9 | 0.0 |
| 16:1n-7 | 2.1 | 20:1n-7 | 1.0 |
| 16:1n-5 | 0.1 | 20:1n-5 | 0.1 |
| 16:2n-4 | 0.2 | 20:2n-9 | 0.0 |
| 17:0 | 0.3 | 20:2n-6 | 0.1 |
| 16:3n-4 | 0.1 | 20:3n-9 | 0.0 |
| 16:3n-3 | 0.0 | 20:3n-6 | 0.0 |
| 16:3n-1 | 0.0 | 20:4n-6 | 0.2 |
| 16:4n-3 | 0.4 | 20:3n-3 | 0.0 |
| 18:0 | 3.2 | 20:4n-3 | 0.1 |
| 18:1n-9 | 32.3 | 20:5n-3 | 2.5 |
| 18:1n-7 | 2.3 | 22:1n-11 | 0.1 |
| 18:1n-5 | 0.0 | 22:1n-9 | 0.3 |
| 18:2n-9 | 0.0 | 22:4n-6 | 0.0 |
| 18:2n-6 | 20.3 | 22:5n-6 | 0.1 |
| 18:2n-4 | 0.1 | 22:5n-3 | 0.3 |
| 18:3n-6 | 0.1 | 22:6n-3 | 1.7 |
| 18:3n-4 | 0.0 |
| Groups | FHD | FLD | VHD | VLD | ANOVA * | ||
|---|---|---|---|---|---|---|---|
| Weight | (kg) | ♀ | 2.40 ± 0.33 | 1.99 ± 0.28 | 2.61 ± 1.19 | 1.84 ± 0.61 | n.s. |
| ♂ | 2.05 ± 0.89 | 1.68 ± 0.13 | 1.30 ± 0.45 | 1.65 ± 0.48 | n.s. | ||
| Lenght | (cm) | ♀ | 45.75 ± 0.35 | 42.83 ± 1.44 | 45.33 ± 5.50 | 41.83 ± 3.75 | n.s. |
| ♂ | 45.50 ± 1.41 | 39.83 ± 1.15 | 36.66 ± 2.02 | 40.83 ± 5.05 | n.s. | ||
| fads2 expression | (fold change) | ♀ | 5.77 ± 2.41a | 0.17 ± 0.18b | 5.29 ± 1.91a | 0.35 ± 0.08b | 0.01 |
| ♂ | 3.05 ± 1.11a | 0.26 ± 0.06b | 6.06 ± 6.26a | 0.72 ± 0.13b | 0.04 | ||
| Main Ingredients (%) | % | Proximate Composition | (% Dry Matter) |
|---|---|---|---|
| Fish meal SA 1 68 super prime | 5.0 | Crude lipids | 21.8 |
| Fish meal alternative protein sources 2 | 54.5 | Crude protein | 57.2 |
| Rapeseed meal cake | 11.3 | Moisture | 6.5 |
| Wheat | 6.9 | Ash | 6.7 |
| Fish oil SA 1 | 3.0 | ||
| Vegetable oil mix 3 | 13.0 | ||
| Micronutrient mixes 4 | 6.3 | Gross Energy (MJ/kg, as is) | 22.5 |
| % Total Fatty Acids | % Total Fatty Acids | ||
| 14:0 | 4.69 | 18:3n-4 | 0.04 |
| 14:1n-5 | 0.17 | 18:3n-3 | 14.86 |
| 15:0 | 0.28 | 18:4n-3 | 0.91 |
| 16:0 | 13.67 | 20:0 | 0.21 |
| 16:1n-7 | 4.50 | 20:1n-9 | 0.37 |
| 16:1n-5 | 0.09 | 20:1n-7 | 7.25 |
| 16:2n-6 | 0.25 | 20:1n-5 | 0.37 |
| 17:0 | 0.13 | 20:2n-6 | 0.16 |
| 16:3n-4 | 0.14 | 20:4n-6 | 0.31 |
| 16:3n-3 | 0.14 | 20:3n-3 | 0.09 |
| 16:3n-1 | 0.04 | 20:4n-3 | 0.19 |
| 16:4n-3 | 0.22 | 20:5n-3 | 3.81 |
| 18:0 | 3.23 | 22:1n-11 | 9.35 |
| 18:1n-9 | 15.75 | 22:1n-9 | 1.00 |
| 18:1n-7 | 2.46 | 22:4n-6 | 0.04 |
| 18:1n-5 | 0.23 | 22:5n-6 | 0.03 |
| 18:2n-6 | 9.84 | 22:5n-3 | 0.28 |
| 18:2n-4 | 0.05 | 22:6n-3 | 4.49 |
| 18:3n-6 | 0.08 |
| Groups | Two-Way ANOVA * | ||||||
|---|---|---|---|---|---|---|---|
| Composition (%) | FHD | FLD | VHD | VLD | Broodstock Diet (D) | Broodstock fads2 Expression (f) | D × f |
| Moisture | 64.2 ± 2.0 | 66.6 ± 5.9 | 66.5 ± 0.1 | 66.6 ± 4.2 | n.s. | n.s. | n.s. |
| Protein | 11.1 ± 0.3 | 10.1 ± 2.4 | 12.1 ± 0.4 | 11.2 ± 1.3 | n.s. | n.s. | n.s. |
| Lipids | 13.9 ± 2.1 | 14.1 ± 4.9 | 12.1 ± 1.8 | 11.0 ± 2.1 | n.s. | n.s. | n.s. |
| Ash | 2.5 ± 0.5 | 2.7 ± 1.2 | 2.9 ± 0.3 | 2.9 ± 0.4 | n.s. | n.s. | n.s. |
| HSI † | 1.4 ± 0.2 | 1.2 ± 0.2 B | 1.3 ± 0.1 b | 1.6 ± 0.1 Aa | n.s. | 0.05 | 0.03 |
| FHD | FLD | VHD | VLD | Diet (D) | fads2(f) | D × f | |
|---|---|---|---|---|---|---|---|
| 14:0 | 1.58 ± 0.09 | 1.66 ± 0.40 | 1.76 ± 0.27 | 1.58 ± 0.28 | 0.76 | 0.76 | 0.47 |
| 14:1n-7 | 0.01 ± 0.00 | 0.01 ± 0.00 | 0.02 ± 0.00 | 0.02 ± 0.00 | 0.14 | 0.69 | 0.52 |
| 14:1n-5 | 0.02 ± 0.01 | 0.04 ± 0.01 | 0.03 ± 0.00 | 0.03 ± 0.00 | 0.80 | 0.20 | 0.16 |
| 15:0 | 0.13 ± 0.01 | 0.17 ± 0.04 | 0.14 ± 0.02 | 0.14 ± 0.00 | 0.68 | 0.12 | 0.20 |
| 15:1n-5 | 0.02 ± 0.00 | 0.02 ± 0.00 | 0.02 ± 0.01 | 0.02 ± 0.00 | 0.88 | 0.51 | 0.11 |
| 16:0ISO | 0.02 ± 0.00B | 0.02 ± 0.00A | 0.02 ± 0.00AB | 0.02 ± 0.00B | 0.49 | 0.43 | 0.03 |
| 16:0 | 15.70 ± 0.42 | 14.30 ± 0.96 | 16.02 ± 1.68 | 13.92 ± 1.92 | 0.97 | 0.06 | 0.67 |
| 16:1n-7 | 2.26 ± 0.17 | 2.49 ± 0.44 | 2.57 ± 0.23 | 2.43 ± 0.23 | 0.48 | 0.79 | 0.31 |
| 16:1n-5 | 0.11 ± 0.00B | 0.13 ± 0.01A | 0.12 ± 0.00B | 0.14 ± 0.02A | 0.24 | 0.04 | 0.38 |
| 16:2n-4 | 0.05 ± 0.02 | 0.09 ± 0.05 | 0.07 ± 0.02 | 0.08 ± 0.01 | 0.71 | 0.11 | 0.37 |
| 17:0 | 0.07 ± 0.01 | 0.11 ± 0.04 | 0.09 ± 0.02 | 0.10 ± 0.01 | 0.52 | 0.09 | 0.27 |
| 16:3n-4 | 0.19 ± 0.01B | 0.21 ± 0.00A | 0.19 ± 0.00B | 0.20 ± 0.00A | 0.80 | 0.00 | 0.07 |
| 16:3n-3 | 0.03 ± 0.00 | 0.04 ± 0.01 | 0.04 ± 0.00 | 0.04 ± 0.01 | 0.67 | 0.08 | 0.08 |
| 16:3n-1 | 0.01 ± 0.00 | 0.01 ± 0.00 | 0.01 ± 0.00 | 0.01 ± 0.00 | 0.11 | 0.50 | 0.19 |
| 16:4n-3 | 0.03 ± 0.01 | 0.05 ± 0.04 | 0.04 ± 0.02 | 0.05 ± 0.01 | 0.63 | 0.17 | 0.46 |
| 18:0 | 6.16 ± 0.54A | 5.38 ± 0.17B | 5.86 ± 0.37AB | 5.40 ± 0.49B | 0.57 | 0.03 | 0.52 |
| 18:1n-9 | 42.01 ± 0.34 | 40.03 ± 2.18 | 40.50 ± 0.48 | 40.05 ± 0.88 | 0.32 | 0.12 | 0.30 |
| 18:1n-7 | 2.52 ± 0.05 | 2.60 ± 0.14 | 2.54 ± 0.07 | 2.56 ± 0.04 | 0.86 | 0.31 | 0.54 |
| 18:1n-5 | 0.06 ± 0.01 | 0.08 ± 0.00 | 0.06 ± 0.03 | 0.07 ± 0.00 | 0.31 | 0.17 | 0.84 |
| 18:2n-9 | 2.04 ± 0.10 | 2.29 ± 0.49 | 2.26 ± 0.33 | 2.69 ± 0.88 | 0.35 | 0.30 | 0.78 |
| 18:2n-6 | 11.09 ± 0.41B | 11.85 ± 0.33A | 10.64 ± 0.56B | 11.67 ± 0.63A | 0.31 | 0.01 | 0.66 |
| 18:2n-4 | 0.06 ± 0.01B | 0.09 ± 0.02A | 0.07 ± 0.00AB | 0.08 ± 0.01AB | 0.85 | 0.01 | 0.26 |
| 18:3n-6 | 1.69 ± 0.04 | 2.17 ± 0.39 | 1.89 ± 0.46 | 2.49 ± 0.85 | 0.41 | 0.11 | 0.85 |
| 18:3n-4 | 0.06 ± 0.00 | 0.07 ± 0.01 | 0.07 ± 0.01 | 0.07 ± 0.00 | 0.32 | 0.20 | 0.11 |
| 18:3n-3 | 5.30 ± 0.24 | 5.67 ± 0.44 | 5.15 ± 0.57 | 5.74 ± 0.45 | 0.89 | 0.10 | 0.68 |
| 18:4n-3 | 1.21 ± 0.01 | 1.60 ± 0.23 | 1.41 ± 0.39 | 1.80 ± 0.53 | 0.36 | 0.09 | 1.00 |
| 18:4n-1 | 0.02 ± 0.00 | 0.04 ± 0.01 | 0.03 ± 0.00 | 0.04 ± 0.00 | 0.32 | 0.10 | 0.19 |
| 20:0 | 0.16 ± 0.00 | 0.15 ± 0.03 | 0.16 ± 0.02 | 0.14 ± 0.02 | 0.69 | 0.41 | 0.87 |
| 20:1n-9 | 0.12 ± 0.01 | 0.14 ± 0.02 | 0.12 ± 0.01 | 0.13 ± 0.01 | 0.51 | 0.19 | 0.69 |
| 20:1n-7 | 0.73 ± 0.05 | 0.71 ± 0.18 | 0.74 ± 0.11 | 0.66 ± 0.09 | 0.83 | 0.48 | 0.69 |
| 20:1n-5 | 0.07 ± 0.00B | 0.08 ± 0.01AB | 0.07 ± 0.01B | 0.08 ± 0.00A | 0.49 | 0.04 | 0.45 |
| 20:2n-9 | 0.75 ± 0.12AB | 0.51 ± 0.12BC | 0.88 ± 0.02A | 0.47 ± 0.14C | 0.52 | 0.00 | 0.23 |
| 20:2n-6 | 0.26 ± 0.02 | 0.25 ± 0.02 | 0.24 ± 0.01 | 0.23 ± 0.02 | 0.11 | 0.23 | 0.92 |
| 20:3n-9 | 0.00 ± 0.00 | 0.01 ± 0.00 | 0.00 ± 0.00 | 0.01 ± 0.00 | 0.71 | 0.12 | 0.48 |
| 20:3n-6 | 0.27 ± 0.03A | 0.22 ± 0.03B | 0.31 ± 0.02A | 0.21 ± 0.04B | 0.37 | 0.00 | 0.19 |
| 20:4n-6 | 0.35 ± 0.00 | 0.39 ± 0.05 | 0.35 ± 0.02 | 0.37 ± 0.02 | 0.57 | 0.17 | 0.50 |
| 20:3n-3 | 0.23 ± 0.02 | 0.22 ± 0.01 | 0.21 ± 0.01 | 0.21 ± 0.01 | 0.07 | 0.46 | 0.67 |
| 20:4n-3 | 0.30 ± 0.03 | 0.32 ± 0.04 | 0.38 ± 0.04 | 0.33 ± 0.01 | 0.07 | 0.39 | 0.12 |
| 20:5n-3 | 0.96 ± 0.12B | 1.42 ± 0.38AB | 1.21 ± 0.10AB | 1.42 ± 0.16A | 0.37 | 0.03 | 0.36 |
| 22:1n-1 | 0.17 ± 0.04 | 0.24 ± 0.12 | 0.21 ± 0.06 | 0.22 ± 0.02 | 0.87 | 0.35 | 0.53 |
| 22:1n-9 | 0.27 ± 0.01 | 0.26 ± 0.04 | 0.26 ± 0.04 | 0.24 ± 0.02 | 0.37 | 0.35 | 0.57 |
| 22:4n-6 | 0.04 ± 0.00 | 0.05 ± 0.01 | 0.05 ± 0.01 | 0.05 ± 0.00 | 0.83 | 0.15 | 0.36 |
| 22:5n-6 | 0.10 ± 0.00 | 0.12 ± 0.01 | 0.10 ± 0.01 | 0.11 ± 0.01 | 0.90 | 0.05 | 0.13 |
| 22:5n-3 | 0.39 ± 0.05B | 0.61 ± 0.12A | 0.49 ± 0.04AB | 0.61 ± 0.09A | 0.35 | 0.01 | 0.37 |
| 22:6n-3 | 2.37 ± 0.16B | 3.09 ± 0.51AB | 2.60 ± 0.18B | 3.10 ± 0.38A | 0.56 | 0.01 | 0.57 |
| Σ | |||||||
| Saturated1 | 23.80 ± 0.84 | 21.77 ± 1.59 | 23.03 ± 1.78 | 21.29 ± 2.58 | 0.57 | 0.11 | 0.89 |
| 16:1n2 | 2.37 ± 0.17 | 2.62 ± 0.45 | 2.60 ± 0.24 | 2.57 ± 0.21 | 0.53 | 0.60 | 0.41 |
| 18:1n | 44.59 ± 0.32 | 42.70 ± 2.04 | 43.53 ± 0.93 | 42.68 ± 0.91 | 0.46 | 0.09 | 0.48 |
| 20:1n | 0.92 ± 0.06 | 0.93 ± 0.21 | 0.91 ± 0.12 | 0.87 ± 0.08 | 0.68 | 0.79 | 0.76 |
| 18:1/16:1 | 18.90 ± 1.52 | 16.70 ± 3.55 | 16.82 ± 1.72 | 16.70 ± 1.31 | 0.87 | 0.65 | 0.67 |
| 100% FO | 30% FO–70% VO | |
|---|---|---|
| Raw Material (%) | (F) | (V) |
| Meals from marine sources 1 | 50.0 | 50.0 |
| Sunflower cake | 13.2 | 13.2 |
| Soya cake 2 | 10.0 | 10.0 |
| Fish oil 3 | 8.0 | 2.4 |
| Linseed oil 4 | - | 5.6 |
| Wheat | 9.9 | 9.9 |
| Corn gluten 60 | 7.0 | 7.0 |
| Drying/wetting | 0.9 | 0.9 |
| Vitamin & mineral premix 5 | 1.0 | 1.0 |
| Vitamin E powder (50%) | 0.1 | 0.1 |
| Biochemical Composition | ||
| (% of Dry Matter) | ||
| Moisture | 9.1 | 8.8 |
| Protein (crude) | 56.3 | 56.1 |
| Lipids (crude) | 17.2 | 17.1 |
| Ash | 8.6 | 8.5 |
| Energy–Gross (MJ kg−1) | 21.2 | 21.2 |
| Fatty Acids (% of Total Fatty Acids) | ||
| 18:2n-6 | 5.6 | 9.9 |
| 18:3n-3 | 0.9 | 16.3 |
| 20:4n-6 | 0.4 | 0.3 |
| 20:5n-3 | 6.3 | 4.8 |
| 22:6n-6 | 7.1 | 6.0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turkmen, S.; Perera, E.; Zamorano, M.J.; Simó-Mirabet, P.; Xu, H.; Pérez-Sánchez, J.; Izquierdo, M. Effects of Dietary Lipid Composition and Fatty Acid Desaturase 2 Expression in Broodstock Gilthead Sea Bream on Lipid Metabolism-Related Genes and Methylation of the fads2 Gene Promoter in Their Offspring. Int. J. Mol. Sci. 2019, 20, 6250. https://doi.org/10.3390/ijms20246250
Turkmen S, Perera E, Zamorano MJ, Simó-Mirabet P, Xu H, Pérez-Sánchez J, Izquierdo M. Effects of Dietary Lipid Composition and Fatty Acid Desaturase 2 Expression in Broodstock Gilthead Sea Bream on Lipid Metabolism-Related Genes and Methylation of the fads2 Gene Promoter in Their Offspring. International Journal of Molecular Sciences. 2019; 20(24):6250. https://doi.org/10.3390/ijms20246250
Chicago/Turabian StyleTurkmen, Serhat, Erick Perera, Maria J. Zamorano, Paula Simó-Mirabet, Hanlin Xu, Jaume Pérez-Sánchez, and Marisol Izquierdo. 2019. "Effects of Dietary Lipid Composition and Fatty Acid Desaturase 2 Expression in Broodstock Gilthead Sea Bream on Lipid Metabolism-Related Genes and Methylation of the fads2 Gene Promoter in Their Offspring" International Journal of Molecular Sciences 20, no. 24: 6250. https://doi.org/10.3390/ijms20246250
APA StyleTurkmen, S., Perera, E., Zamorano, M. J., Simó-Mirabet, P., Xu, H., Pérez-Sánchez, J., & Izquierdo, M. (2019). Effects of Dietary Lipid Composition and Fatty Acid Desaturase 2 Expression in Broodstock Gilthead Sea Bream on Lipid Metabolism-Related Genes and Methylation of the fads2 Gene Promoter in Their Offspring. International Journal of Molecular Sciences, 20(24), 6250. https://doi.org/10.3390/ijms20246250