Increased Cytotoxicity of Herpes Simplex Virus Thymidine Kinase Expression in Human Induced Pluripotent Stem Cells
Abstract
:1. Introduction
2. Results
2.1. Human iPSCs Transduced with Lentiviral Vectors Expressing HSV-TK
2.2. Targeted Insertion of the HSV-TK Gene into the GAPDH Locus with CRISPR/Cas9-Mediated Genome Editing
2.3. Cytotoxicity of HSV-TK Expression Investigated with Tet-Inducible Lentiviral Vectors
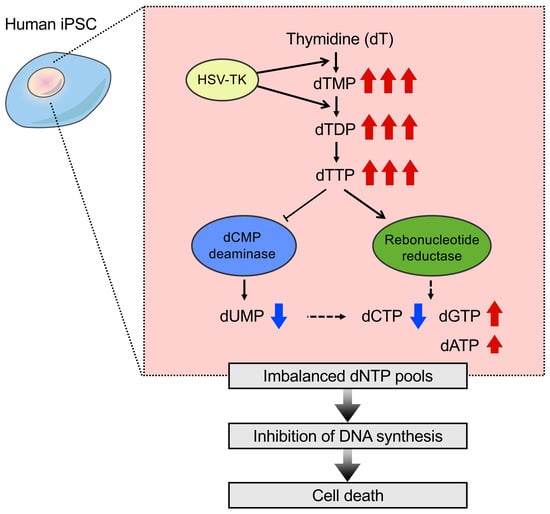
2.4. Nucleotide Metabolism in iPSCs That Expressed HSV-TK
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Lentiviral Vector Preparation
4.3. CRISPR/Cas9-Mediated Genome Editing
- GAPDH-GF1, 5′-CTTCTCTGCTGTAGGCTCATTTG-3′;
- GAPDH-GR1, 5′- AACTCCTGACCTCAGGTGATACA-3′;
- GAPDH-F1, 5′-CTAGGTATGACAACGAATTTGGC-3′;
- GAPDH-R1, 5′-TGGTTGAGCACAGGGTACTTTAT-3′;
- HSV1tk-R1, 5′-GTCTTAGGGCAGTTCTCCTGTTG-3′;
- Puro-GF1, 5′-GAGCTGCAAGAACTCTTCCTCAC-3′;
- Venus-GF1, 5′-ACAACCACTACCTGAGCTACCAG-3′;
- Venus-GR1, 5′-GTAGTTGTACTCCAGCTTGTGCC-3′.
4.4. Cell Viability Assay
4.5. Metabolome Analysis
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| iPSC | induced pluripotent stem cell |
| ESC | embryonic stem cell |
| NS/PC | neural stem/progenitor cell |
| HSV-TK | herpes simplex virus type 1 thymidine kinase |
| GCV | gancicrovir |
| EF-1α | elongation factor 1 α subunit |
| MOI | multiplicity of infection |
| hKO1 | humanized-codon Kusabira-Orange |
| FACS | fluorescence-activated cell sorting |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| HR | homologous recombination |
| Tet | tetracycline |
| GFP | green fluorescent protein |
| Dox | doxycycline |
| dT | thymidine |
| dU | deoxyuridine |
| dC | deoxycytidine |
| dTMP | thymidine monophosphate |
| dTDP | thymidine diphosphate |
| dTTP | thymidine triphosphate |
| CD | dCMP deaminase |
| RNR | ribonucleotide reductase |
| dNTP | deoxyribonucleoside triphosphate |
| NTP | ribonucleoside triphosphate |
| PGK | phosphoglycerate kinase |
| OXPHOS | oxidative phosphorylation |
| CCK-8 | Cell Counting Kit-8 |
| WST-8 | water-soluble tetrazolium salt |
| IC | ion-chromatography |
References
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef] [PubMed]
- Okano, H.; Yamanaka, S. iPS cell technologies: significance and applications to CNS regeneration and disease. Mol. Brain 2014, 7, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Inoue, H.; Wu, J.C.; Yamanaka, S. Induced pluripotent stem cell technology: a decade of progress. Nat. Rev. Drug Discov. 2017, 16, 115–130. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, O.; Sugai, K.; Yamaguchi, R.; Tashiro, S.; Nagoshi, N.; Kohyama, J.; Iida, T.; Ohkubo, T.; Itakura, G.; Isoda, M.; et al. Concise Review: Laying the Groundwork for a First-In-Human Study of an Induced Pluripotent Stem Cell-Based Intervention for Spinal Cord Injury. Stem Cells 2018. [Google Scholar] [CrossRef] [PubMed]
- Ben-David, U.; Benvenisty, N. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat. Rev. Cancer 2011, 11, 268–277. [Google Scholar] [CrossRef] [PubMed]
- Lund, R.J.; Narva, E.; Lahesmaa, R. Genetic and epigenetic stability of human pluripotent stem cells. Nat. Rev. Genet. 2012, 13, 732–744. [Google Scholar] [CrossRef]
- Okano, H.; Nakamura, M.; Yoshida, K.; Okada, Y.; Tsuji, O.; Nori, S.; Ikeda, E.; Yamanaka, S.; Miura, K. Steps Toward Safe Cell Therapy Using Induced Pluripotent Stem Cells. Circ. Res. 2013, 112, 523–533. [Google Scholar] [CrossRef] [Green Version]
- Nori, S.; Okada, Y.; Nishimura, S.; Sasaki, T.; Itakura, G.; Kobayashi, Y.; Renault-Mihara, F.; Shimizu, A.; Koya, I.; Yoshida, R.; et al. Long-term safety issues of iPSC-based cell therapy in a spinal cord injury model: Oncogenic transformation with epithelial-mesenchymal transition. Stem Cell Rep. 2015, 4, 360–373. [Google Scholar] [CrossRef]
- Tang, C.; Lee, A.S.; Volkmer, J.P.; Sahoo, D.; Nag, D.; Mosley, A.R.; Inlay, M.A.; Ardehali, R.; Chavez, S.L.; Pera, R.R.; et al. An antibody against SSEA-5 glycan on human pluripotent stem cells enables removal of teratoma-forming cells. Nat. Biotechnol. 2011, 29, 829–834. [Google Scholar] [CrossRef] [Green Version]
- Tohyama, S.; Hattori, F.; Sano, M.; Hishiki, T.; Nagahata, Y.; Matsuura, T.; Hashimoto, H.; Suzuki, T.; Yamashita, H.; Satoh, Y.; et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell 2013, 12, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.O.; Moon, S.H.; Jeong, H.C.; Yi, J.Y.; Lee, T.H.; Shim, S.H.; Rhee, Y.H.; Lee, S.H.; Oh, S.J.; Lee, M.Y.; et al. Inhibition of pluripotent stem cell-derived teratoma formation by small molecules. Proc. Natl. Acad. Sci. USA 2013, 110, E3281–E3290. [Google Scholar] [CrossRef] [PubMed]
- Ben-David, U.; Gan, Q.F.; Golan-Lev, T.; Arora, P.; Yanuka, O.; Oren, Y.S.; Leikin-Frenkel, A.; Graf, M.; Garippa, R.; Boehringer, M.; et al. Selective elimination of human pluripotent stem cells by an oleate synthesis inhibitor discovered in a high-throughput screen. Cell. Stem Cell 2013, 12, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Okubo, T.; Iwanami, A.; Kohyama, J.; Itakura, G.; Kawabata, S.; Nishiyama, Y.; Sugai, K.; Ozaki, M.; Iida, T.; Matsubayashi, K.; et al. Pretreatment with a gamma-Secretase Inhibitor Prevents Tumor-like Overgrowth in Human iPSC-Derived Transplants for Spinal Cord Injury. Stem Cell Rep. 2016, 7, 649–663. [Google Scholar] [CrossRef] [PubMed]
- Kiuru, M.; Boyer, J.L.; O’Connor, T.P.; Crystal, R.G. Genetic control of wayward pluripotent stem cells and their progeny after transplantation. Cell Stem Cell 2009, 4, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Ke, Q.; Chen, F.; Cai, B.; Gao, Y.; Ye, C.; Wang, D.; Zhang, L.; Lahn, B.T.; Li, W.; et al. Protecting against wayward human induced pluripotent stem cells with a suicide gene. Biomaterials 2012, 33, 3195–3204. [Google Scholar] [CrossRef]
- Yagyu, S.; Hoyos, V.; Del Bufalo, F.; Brenner, M.K. An Inducible Caspase-9 Suicide Gene to Improve the Safety of Therapy Using Human Induced Pluripotent Stem Cells. Mol. Ther. 2015, 23, 1475–1485. [Google Scholar] [CrossRef]
- Itakura, G.; Kawabata, S.; Ando, M.; Nishiyama, Y.; Sugai, K.; Ozaki, M.; Iida, T.; Ookubo, T.; Kojima, K.; Kashiwagi, R.; et al. Fail-Safe System against Potential Tumorigenicity after Transplantation of iPSC Derivatives. Stem Cell Rep. 2017, 8, 673–684. [Google Scholar] [CrossRef]
- Kojima, K.; Miyoshi, H.; Nagoshi, N.; Kohyama, J.; Itakura, G.; Kawabata, S.; Ozaki, M.; Iida, T.; Sugai, K.; Ito, S.; et al. Selective Ablation of Tumorigenic Cells Following Human Induced Pluripotent Stem Cell-Derived Neural Stem/Progenitor Cell Transplantation in Spinal Cord Injury. Stem Cells Transl. Med. in press. [CrossRef]
- Gentry, G.A. Viral thymidine kinases and their relatives. Pharmacol. Ther. 1992, 54, 319–355. [Google Scholar] [CrossRef]
- Fillat, C.; Carrio, M.; Cascante, A.; Sangro, B. Suicide gene therapy mediated by the Herpes Simplex virus thymidine kinase gene/Ganciclovir system: fifteen years of application. Curr. Gene Ther. 2003, 3, 13–26. [Google Scholar] [CrossRef]
- van Dillen, I.J.; Mulder, N.H.; Vaalburg, W.; de Vries, E.F.; Hospers, G.A. Influence of the bystander effect on HSV-tk/GCV gene therapy. A review. Curr. Gene Ther. 2002, 2, 307–322. [Google Scholar] [CrossRef] [PubMed]
- Dachs, G.U.; Hunt, M.A.; Syddall, S.; Singleton, D.C.; Patterson, A.V. Bystander or no bystander for gene directed enzyme prodrug therapy. Molecules 2009, 14, 4517–4545. [Google Scholar] [CrossRef] [PubMed]
- Cihova, M.; Altanerova, V.; Altaner, C. Stem cell based cancer gene therapy. Mol. Pharm. 2011, 8, 1480–1487. [Google Scholar] [CrossRef]
- Okura, H.; Smith, C.A.; Rutka, J.T. Gene therapy for malignant glioma. Mol. Cell Ther. 2014, 2, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stuckey, D.W.; Shah, K. Stem cell-based therapies for cancer treatment: Separating hope from hype. Nat. Rev. Cancer 2014, 14, 683–691. [Google Scholar] [CrossRef]
- Nagai, T.; Ibata, K.; Park, E.S.; Kubota, M.; Mikoshiba, K.; Miyawaki, A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002, 20, 87–90. [Google Scholar] [CrossRef]
- Karasawa, S.; Araki, T.; Nagai, T.; Mizuno, H.; Miyawaki, A. Cyan-emitting and orange-emitting fluorescent proteins as a donor/acceptor pair for fluorescence resonance energy transfer. Biochem. J. 2004, 381, 307–312. [Google Scholar] [CrossRef]
- Al-Shawi, R.; Burke, J.; Wallace, H.; Jones, C.; Harrison, S.; Buxton, D.; Maley, S.; Chandley, A.; Bishop, J.O. The herpes simplex virus type 1 thymidine kinase is expressed in the testes of transgenic mice under the control of a cryptic promoter. Mol. Cell Biol. 1991, 11, 4207–4216. [Google Scholar] [CrossRef]
- Cai, L.Y.; Kato, T.; Ito, K.; Nakayama, M.; Susa, T.; Aikawa, S.; Maeda, K.; Tsukamura, H.; Ohta, A.; Izumi, S.; et al. Expression of porcine FSHbeta subunit promoter-driven herpes simplex virus thymidine kinase gene in transgenic rats. J. Reprod. Dev. 2007, 53, 201–209. [Google Scholar] [CrossRef]
- Cai, L.Y.; Kato, T.; Nakayama, M.; Susa, T.; Murakami, S.; Izumi, S.; Kato, Y. HSV type 1 thymidine kinase protein accumulation in round spermatids induces male infertility by spermatogenesis disruption and apoptotic loss of germ cells. Reprod. Toxicol. 2009, 27, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Salomon, B.; Maury, S.; Loubiere, L.; Caruso, M.; Onclercq, R.; Klatzmann, D. A truncated herpes simplex virus thymidine kinase phosphorylates thymidine and nucleoside analogs and does not cause sterility in transgenic mice. Mol. Cell Biol. 1995, 15, 5322–5328. [Google Scholar] [CrossRef] [PubMed]
- Nordlund, P.; Reichard, P. Ribonucleotide reductases. Annu. Rev. Biochem. 2006, 75, 681–706. [Google Scholar] [CrossRef]
- Hofer, A.; Crona, M.; Logan, D.T.; Sjoberg, B.M. DNA building blocks: Keeping control of manufacture. Crit. Rev. Biochem. Mol. Biol. 2012, 47, 50–63. [Google Scholar] [CrossRef]
- Bjursell, G.; Reichard, P. Effects of thymidine on deoxyribonucleoside triphosphate pools and deoxyribonucleic acid synthesis in Chinese hamster ovary cells. J. Biol. Chem. 1973, 248, 3904–3909. [Google Scholar] [PubMed]
- Schuldiner, M.; Itskovitz-Eldor, J.; Benvenisty, N. Selective ablation of human embryonic stem cells expressing a “suicide” gene. Stem Cells 2003, 21, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Tieng, V.; Cherpin, O.; Gutzwiller, E.; Zambon, A.C.; Delgado, C.; Salmon, P.; Dubois-Dauphin, M.; Krause, K.H. Elimination of proliferating cells from CNS grafts using a Ki67 promoter-driven thymidine kinase. Mol. Ther. Methods Clin. Dev. 2016, 6, 16069. [Google Scholar] [CrossRef]
- Cho, Y.M.; Kwon, S.; Pak, Y.K.; Seol, H.W.; Choi, Y.M.; Park, D.J.; Park, K.S.; Lee, H.K. Dynamic changes in mitochondrial biogenesis and antioxidant enzymes during the spontaneous differentiation of human embryonic stem cells. Biochem. Biophys. Res. Commun. 2006, 348, 1472–1478. [Google Scholar] [CrossRef]
- Varum, S.; Rodrigues, A.S.; Moura, M.B.; Momcilovic, O.; Easley, C.A.t.; Ramalho-Santos, J.; Van Houten, B.; Schatten, G. Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS ONE 2011, 6, e20914. [Google Scholar] [CrossRef]
- Folmes, C.D.; Terzic, A. Energy metabolism in the acquisition and maintenance of stemness. Semin. Cell Dev. Biol. 2016, 52, 68–75. [Google Scholar] [CrossRef] [Green Version]
- Reichard, P. Interactions between deoxyribonucleotide and DNA synthesis. Annu. Rev. Biochem. 1988, 57, 349–374. [Google Scholar] [CrossRef] [PubMed]
- Fairman, J.W.; Wijerathna, S.R.; Ahmad, M.F.; Xu, H.; Nakano, R.; Jha, S.; Prendergast, J.; Welin, R.M.; Flodin, S.; Roos, A.; et al. Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization. Nat. Struct. Mol. Biol 2011, 18, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Boward, B.; Wu, T.; Dalton, S. Concise Review: Control of Cell Fate Through Cell Cycle and Pluripotency Networks. Stem Cells 2016, 34, 1427–1436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wanet, A.; Arnould, T.; Najimi, M.; Renard, P. Connecting Mitochondria, Metabolism, and Stem Cell Fate. Stem Cells Dev. 2015, 24, 1957–1971. [Google Scholar] [CrossRef] [PubMed]
- Weinberger, L.; Ayyash, M.; Novershtern, N.; Hanna, J.H. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol. 2016, 17, 155–169. [Google Scholar] [CrossRef] [PubMed]
- Teslaa, T.; Teitell, M.A. Pluripotent stem cell energy metabolism: An update. EMBO J. 2015, 34, 138–153. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, J.; Ruohola-Baker, H. Metabolic remodeling during the loss and acquisition of pluripotency. Development 2017, 144, 541–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herst, P.M.; Berridge, M.V. Cell surface oxygen consumption: A major contributor to cellular oxygen consumption in glycolytic cancer cell lines. Biochim. Biophys. Acta. 2007, 1767, 170–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jose, C.; Bellance, N.; Rossignol, R. Choosing between glycolysis and oxidative phosphorylation: A tumor’s dilemma? Biochim. Biophys. Acta. 2011, 1807, 552–561. [Google Scholar] [CrossRef]
- Reitzer, L.J.; Wice, B.M.; Kennell, D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J. Biol. Chem. 1979, 254, 2669–2676. [Google Scholar]
- Rodriguez-Enriquez, S.; Vital-Gonzalez, P.A.; Flores-Rodriguez, F.L.; Marin-Hernandez, A.; Ruiz-Azuara, L.; Moreno-Sanchez, R. Control of cellular proliferation by modulation of oxidative phosphorylation in human and rodent fast-growing tumor cells. Toxicol. Appl. Pharmacol. 2006, 215, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, M.; Koyanagi, M.; Tanabe, K.; Takahashi, K.; Ichisaka, T.; Aoi, T.; Okita, K.; Mochiduki, Y.; Takizawa, N.; Yamanaka, S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 2008, 26, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, M.; Taniguchi, Y.; Senda, S.; Takizawa, N.; Ichisaka, T.; Asano, K.; Morizane, A.; Doi, D.; Takahashi, J.; Nishizawa, M.; et al. A novel efficient feeder-free culture system for the derivation of human induced pluripotent stem cells. Sci. Rep. 2014, 4, 3594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozaki, M.; Iwanami, A.; Nagoshi, N.; Kohyama, J.; Itakura, G.; Iwai, H.; Nishimura, S.; Nishiyama, Y.; Kawabata, S.; Sugai, K.; et al. Evaluation of the immunogenicity of human iPS cell-derived neural stem/progenitor cells in vitro. Stem Cell Res. 2017, 19, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Yusa, K.; Rashid, S.T.; Strick-Marchand, H.; Varela, I.; Liu, P.Q.; Paschon, D.E.; Miranda, E.; Ordonez, A.; Hannan, N.R.; Rouhani, F.J.; et al. Targeted gene correction of alpha1-antitrypsin deficiency in induced pluripotent stem cells. Nature 2011, 478, 391–394. [Google Scholar] [CrossRef]
- Iida, T.; Iwanami, A.; Sanosaka, T.; Kohyama, J.; Miyoshi, H.; Nagoshi, N.; Kashiwagi, R.; Toyama, Y.; Matsumoto, M.; Nakamura, M.; et al. Whole-Genome DNA Methylation Analyses Revealed Epigenetic Instability in Tumorigenic Human iPS Cell-Derived Neural Stem/Progenitor Cells. Stem Cells 2017, 35, 1316–1327. [Google Scholar] [CrossRef]
- Hashizume, O.; Ohnishi, S.; Mito, T.; Shimizu, A.; Ishikawa, K.; Nakada, K.; Soda, M.; Mano, H.; Togayachi, S.; Miyoshi, H.; et al. Epigenetic regulation of the nuclear-coded GCAT and SHMT2 genes confers human age-associated mitochondrial respiration defects. Sci. Rep. 2015, 5, 10434. [Google Scholar] [CrossRef] [Green Version]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.L.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.B.; Jiang, W.Y.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [Green Version]
- Semba, H.; Takeda, N.; Isagawa, T.; Sugiura, Y.; Honda, K.; Wake, M.; Miyazawa, H.; Yamaguchi, Y.; Miura, M.; Jenkins, D.M.; et al. HIF-1alpha-PDK1 axis-induced active glycolysis plays an essential role in macrophage migratory capacity. Nat. Commun. 2016, 7, 11635. [Google Scholar] [CrossRef]




| Human iPSCs | No. of FACS-Sorted Venus-Positive Cells/Well | ||
|---|---|---|---|
| 1 | 10 | 100 | |
| (No. of Surviving Cells/Total No. of Sorted Cells) | |||
| 253G1 | 0/26 | 0/10 | 1*/100 |
| 1231A3 | 1*/26 | 0/10 | 1*/100 |
| 1210B2 | 0/26 | 0/10 | 1*/200 |
| 1210B2 (Cont.) | 6/24 | 2/20 | 20/200 |
| Human Cells | No. of Puror Colonies | |
|---|---|---|
| Exp. 1 | Exp. 2 | |
| 253G1 | 1 | 0 |
| 1231A3 | 1 | 0 |
| 1210B2 | 1 | 2 |
| HeLa | >20 | ND |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iwasawa, C.; Tamura, R.; Sugiura, Y.; Suzuki, S.; Kuzumaki, N.; Narita, M.; Suematsu, M.; Nakamura, M.; Yoshida, K.; Toda, M.; et al. Increased Cytotoxicity of Herpes Simplex Virus Thymidine Kinase Expression in Human Induced Pluripotent Stem Cells. Int. J. Mol. Sci. 2019, 20, 810. https://doi.org/10.3390/ijms20040810
Iwasawa C, Tamura R, Sugiura Y, Suzuki S, Kuzumaki N, Narita M, Suematsu M, Nakamura M, Yoshida K, Toda M, et al. Increased Cytotoxicity of Herpes Simplex Virus Thymidine Kinase Expression in Human Induced Pluripotent Stem Cells. International Journal of Molecular Sciences. 2019; 20(4):810. https://doi.org/10.3390/ijms20040810
Chicago/Turabian StyleIwasawa, Chizuru, Ryota Tamura, Yuki Sugiura, Sadafumi Suzuki, Naoko Kuzumaki, Minoru Narita, Makoto Suematsu, Masaya Nakamura, Kazunari Yoshida, Masahiro Toda, and et al. 2019. "Increased Cytotoxicity of Herpes Simplex Virus Thymidine Kinase Expression in Human Induced Pluripotent Stem Cells" International Journal of Molecular Sciences 20, no. 4: 810. https://doi.org/10.3390/ijms20040810
APA StyleIwasawa, C., Tamura, R., Sugiura, Y., Suzuki, S., Kuzumaki, N., Narita, M., Suematsu, M., Nakamura, M., Yoshida, K., Toda, M., Okano, H., & Miyoshi, H. (2019). Increased Cytotoxicity of Herpes Simplex Virus Thymidine Kinase Expression in Human Induced Pluripotent Stem Cells. International Journal of Molecular Sciences, 20(4), 810. https://doi.org/10.3390/ijms20040810