Changes in the Soil Microbiome in Eggplant Monoculture Revealed by High-Throughput Illumina MiSeq Sequencing as Influenced by Raw Garlic Stalk Amendment
Abstract
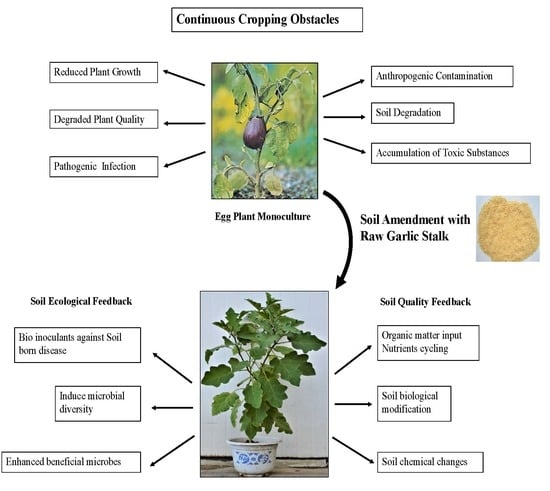
:1. Introduction
2. Results
2.1. Effect of RGS on Soil Chemical Properties
2.2. Soil Microbe Identifications
2.3. Soil Microbial Diversity
2.4. Venn Diagram Analysis
2.5. Changes in Bacterial and Fungal Community
2.6. Linear Discriminant Analysis Effect Size (LEfSE) Analysis of Microbial Community
2.7. Top Abundant Microbial Distribution Revealed by Heat Map Analysis
2.8. Principal Component Analysis (PCA)
2.9. Hierarchical Cluster Analysis of Fungal Community in RGS Treatments
2.10. Correlation between Soil Chemical Characteristics and Soil Microbial Community
3. Discussion
3.1. Effect of RGS on Soil Chemical Properties
3.2. Effect of RGS on Soil Microbial Communities
3.2.1. Bacterial Abundance and Community Structure
3.2.2. Fungi Abundance and Community Structure
4. Materials and Methods
4.1. Experiemtnal Description and Soil Sampling
4.2. Soil Chemical Analyses
4.3. Assessment of Soil Enzyme Activities
4.4. Microbial DNA Extraction and PCR Amplification
4.5. Profiling of Illumina MiSeq Analyses
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zhang, A.; Cheng, G.; Hussain, Q.; Zhang, M.; Feng, H.; Dyck, M.; Sun, B.; Zhao, Y.; Chen, H.; Chen, J. Contrasting effects of straw and straw–derived biochar application on net global warming potential in the Loess Plateau of China. Field Crop. Res. 2017, 205, 45–54. [Google Scholar] [CrossRef]
- Bailey, K.; Lazarovits, G. Suppressing soil-borne diseases with residue management and organic amendments. Soil Tillage Res. 2003, 72, 169–180. [Google Scholar] [CrossRef]
- Sial, T.A.; Khan, M.N.; Lan, Z.; Kumbhar, F.; Zhao, Y.; Zhang, J.; Sun, D.; Xiu, L. Contrasting effects of banana peels waste and its biochar on greenhouse gas emissions and soil biochemical properties. Process Saf. Environ. Prot. 2018. [Google Scholar] [CrossRef]
- Sial, T.A.; Liu, J.; Zhao, Y.; Khan, M.N.; Lan, Z.; Zhang, J.; Kumbhar, F.; Akhtar, K.; Rajpar, I. Co-Application of Milk Tea Waste and NPK Fertilizers to Improve Sandy Soil Biochemical Properties and Wheat Growth. Molecules 2019, 24, 423. [Google Scholar] [CrossRef]
- Bhattacharjya, S.; Chandra, R.; Sharma, M.P.; Sharma, S.K.; Agnihotri, R. Biochar and Crop Residue Amendments on Soil Microbial and Biochemical Properties. Proc. Natl. Acad. Sci. India Sect. B 2017, 87, 975–983. [Google Scholar] [CrossRef]
- Li, T.; Liu, T.; Zheng, C.; Kang, C.; Yang, Z.; Yao, X.; Song, F.; Zhang, R.; Wang, X.; Xu, N. Changes in soil bacterial community structure as a result of incorporation of Brassica plants compared with continuous planting eggplant and chemical disinfection in greenhouses. PloS ONE 2017, 12, e0173923. [Google Scholar] [CrossRef] [PubMed]
- Mihajlović, M.; Rekanović, E.; Hrustić, J.; Grahovac, M.; Tanović, B. Methods for management of soilborne plant pathogens. Pestic. I Fitomedicina 2017, 32, 9–24. [Google Scholar] [CrossRef]
- Faostat, F. Agriculture Organization of the United Nations Statistics Division. Economic and Social Development Department, Rome, Italy. Available online: http://faostat3. fao. org/home/E (accessed on 31 December 2016).
- Lee, Y.; Kim, Y.; Oh, Y.; Ahmadi, F.; Kwak, W. Yield survey and nutritional evaluation of garlic stalk for ruminant feed. J. Anim. Sci. Technol. 2017, 59, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghani, M.I.; Ali, A.; Atif, M.J.; Ali, M.; Amin, B.; Anees, M.; Cheng, Z. Soil Amendment with Raw Garlic Stalk: A Novel Strategy to Stimulate Growth and the Antioxidative Defense System in Monocropped Eggplant in the North of China. Agronomy 2019, 9, 89. [Google Scholar] [CrossRef]
- Auger, J. Insecticidal and fungicidal potential of Allium substances as biofumigants. Agondustria 2004, 3, 5–8. [Google Scholar]
- Zhang, M.; Cheng, G.; Feng, H.; Sun, B.; Zhao, Y.; Chen, H.; Chen, J.; Dyck, M.; Wang, X.; Zhang, J. Effects of straw and biochar amendments on aggregate stability, soil organic carbon, and enzyme activities in the Loess Plateau, China. Environ. Sci. Pollut. Res. 2017, 24, 10108–10120. [Google Scholar] [CrossRef]
- Chen, L.-H.; Han, R.; Zhang, H.; Xu, X.-H.; Shao, H.-B.; Wang, M.-Y.; Cheng, Y.; Shao, X.-H. Irrigating-continuous cropping with Bacillus subtilis D9 fortified waste water could control the Fusarium wilt of Artemisia selengens. Appl. Soil Ecol. 2017, 113, 127–134. [Google Scholar] [CrossRef]
- Xie, X.-G.; Dai, C.-C.; Li, X.-G.; Wu, J.-R.; Wu, Q.-Q.; Wang, X.-X. Reduction of soil-borne pathogen Fusarium solani reproduction in soil enriched with phenolic acids by inoculation of endophytic fungus Phomopsis liquidambari. BioControl 2017, 62, 111–123. [Google Scholar] [CrossRef]
- Xiao, X.; Cheng, Z.; Meng, H.; Khan, M.A.; Li, H. Intercropping with garlic alleviated continuous cropping obstacle of cucumber in plastic tunnel. Acta Agric. Scand. Sect. B Soil Plant Sci. 2012, 62, 696–705. [Google Scholar] [CrossRef]
- Li, H.; Zhang, L.; Jiang, X.; Liu, Q. Allelopathic effects of phenolic acids on the growth and physiological characteristics of strawberry plants. Allelopath. J. 2015, 35, 61–75. [Google Scholar]
- Xin-hui, Z.; Lang, D.-y.; Zhang, E.-h.; Wang, Z.-s. Effect of autotoxicity and soil microbes in continuous cropping soil on angelica sinensis seedling growth and rhizosphere soil microbial population. Chin. Herb. Med. 2015, 7, 88–93. [Google Scholar] [CrossRef]
- Yao, H.; Jiao, X.; Wu, F. Effects of continuous cucumber cropping and alternative rotations under protected cultivation on soil microbial community diversity. Plant Soil 2006, 284, 195–203. [Google Scholar] [CrossRef]
- Wu, H.; Pratley, J.; Lemerle, D.; An, M.; Li Liu, D. Autotoxicity of wheat (Triticum aestivum L.) as determined by laboratory bioassays. Plant Soil 2007, 296, 85–93. [Google Scholar] [CrossRef]
- Ding, H.; Ali, A.; Cheng, Z. Dynamics of a Soil Fungal Community in a Three-Year Green Garlic/Cucumber Crop Rotation System in Northwest China. Sustainability (2071-1050) 2018, 10, 1391. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, E.; Wang, H.; Lang, D. Effects of continuous cropping obstacle on growth of Angelica sinensis and its mechanism. China J. Chin. Mater. Med. 2010, 35, 1231–1234. [Google Scholar]
- Al-Saleh, M. Evaluation of saudi fluorescent Pseudomonads isolates as a biocontrol agent against citrus canker disease caused by Xanthomonas citri subsp citri a. Egypt Acad. J. Biol. Sci. 2014, 6, 1–7. [Google Scholar] [CrossRef]
- Han, X.; Cheng, Z.; Meng, H. Soil properties, nutrient dynamics, and soil enzyme activities associated with garlic stalk decomposition under various conditions. PloS ONE 2012, 7, e50868. [Google Scholar] [CrossRef] [PubMed]
- Kabirinejad, S.; Kalbasi, M.; Khoshgoftarmanesh, A.H.; Hoodaji, M.; Afyuni, M. Effect of incorporation of crops residue into soil on some chemical properties of soil and bioavailability of copper in soil. Int. J. Adv. Biol. Biomed. Res. 2014, 2, 2819–2824. [Google Scholar]
- Eskelinen, A.; Stark, S.; Männistö, M. Links between plant community composition, soil organic matter quality and microbial communities in contrasting tundra habitats. Oecologia 2009, 161, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Blagodatskaya, Е.; Kuzyakov, Y. Mechanisms of real and apparent priming effects and their dependence on soil microbial biomass and community structure: Critical review. Biol. Fertil. Soils 2008, 45, 115–131. [Google Scholar] [CrossRef]
- Jeong, C.Y.; Dodla, S.K.; Wang, J.J. Fundamental and molecular composition characteristics of biochars produced from sugarcane and rice crop residues and by-products. Chemosphere 2016, 142, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.-L.; Fan, L.-L.; Xie, G.-J. Effect of copper on the performance and bacterial communities of activated sludge using Illumina MiSeq platforms. Chemosphere 2016, 156, 212–219. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, A.; Bhadury, P. Insights into bacterioplankton community structure from Sundarbans mangrove ecoregion using Sanger and Illumina MiSeq sequencing approaches: A comparative analysis. Genom. Data 2017, 11, 39–42. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Chen, L.; Zhang, R.; Lin, K. High throughput sequencing analysis of the joint effects of BDE209-Pb on soil bacterial community structure. J. Hazard. Mater. 2016, 301, 1–7. [Google Scholar] [CrossRef]
- Wu, L.; Chen, J.; Xiao, Z.; Zhu, X.; Wang, J.; Wu, H.; Wu, Y.; Zhang, Z.; Lin, W. Barcoded Pyrosequencing Reveals a Shift in the Bacterial Community in the Rhizosphere and Rhizoplane of Rehmannia glutinosa under Consecutive Monoculture. Int. J. Mol. Sci. 2018, 19, 850. [Google Scholar] [CrossRef]
- Wang, W.; Wang, H.; Feng, Y.; Wang, L.; Xiao, X.; Xi, Y.; Luo, X.; Sun, R.; Ye, X.; Huang, Y. Consistent responses of the microbial community structure to organic farming along the middle and lower reaches of the Yangtze River. Sci. Rep. 2016, 6, 35046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiomi, Y.; Nishiyama, M.; Onizuka, T.; Marumoto, T. Comparison of bacterial community structures in the rhizoplane of tomato plants grown in soils suppressive and conducive towards bacterial wilt. Appl. Environ. Microbiol. 1999, 65, 3996–4001. [Google Scholar] [PubMed]
- Jiang, J.; Song, Z.; Yang, X.; Mao, Z.; Nie, X.; Guo, H.; Peng, X. Microbial community analysis of apple rhizosphere around Bohai Gulf. Sci. Rep. 2017, 7, 8918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trivedi, P.; Anderson, I.C.; Singh, B.K. Microbial modulators of soil carbon storage: Integrating genomic and metabolic knowledge for global prediction. Trends Microbiol. 2013, 21, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Cornejo, N.L.; Romero-Salas, E.A.; Navarro-Noya, Y.E.; González-Zúñiga, J.C.; Ramirez-Villanueva, D.A.; Vásquez-Murrieta, M.S.; Verhulst, N.; Govaerts, B.; Dendooven, L.; Luna-Guido, M. Incorporation of bean plant residue in soil with different agricultural practices and its effect on the soil bacteria. Appl. Soil Ecol. 2017, 119, 417–427. [Google Scholar] [CrossRef]
- Lueders, T.; Kindler, R.; Miltner, A.; Friedrich, M.W.; Kaestner, M. Identification of bacterial micropredators distinctively active in a soil microbial food web. Appl. Environ. Microbiol. 2006, 72, 5342–5348. [Google Scholar] [CrossRef] [PubMed]
- Hervé, V.; Le Roux, X.; Uroz, S.; Gelhaye, E.; Frey-Klett, P. Diversity and structure of bacterial communities associated with P hanerochaete chrysosporium during wood decay. Environ. Microbiol. 2014, 16, 2238–2252. [Google Scholar] [CrossRef]
- Folman, L.B.; Klein Gunnewiek, P.J.; Boddy, L.; De Boer, W. Impact of white-rot fungi on numbers and community composition of bacteria colonizing beech wood from forest soil. FEMS Microbiol. Ecol. 2008, 63, 181–191. [Google Scholar] [CrossRef] [Green Version]
- Visioli, G.; Sanangelantoni, A.; Vamerali, T.; Dal Cortivo, C.; Blandino, M. 16S rDNA Profiling to Reveal the Influence of Seed-Applied Biostimulants on the Rhizosphere of Young Maize Plants. Molecules 2018, 23, 1461. [Google Scholar] [CrossRef]
- Park, S.; Yu, J.; Byun, I.; Cho, S.; Park, T.; Lee, T. Microbial community structure and dynamics in a mixotrophic nitrogen removal process using recycled spent caustic under different loading conditions. Bioresour. Technol. 2011, 102, 7265–7271. [Google Scholar] [CrossRef]
- McBride, M.J.; Liu, W.; Lu, X.; Zhu, Y.; Zhang, W. The family cytophagaceae. In The Prokaryotes; Springer: Berlin/Heidelberg, Germany, 2014; pp. 577–593. [Google Scholar]
- Hayat, R.; Ali, S.; Amara, U.; Khalid, R.; Ahmed, I. Soil beneficial bacteria and their role in plant growth promotion: A review. Ann. Microbiol. 2010, 60, 579–598. [Google Scholar] [CrossRef]
- Zheng, J.; Chen, J.; Pan, G.; Wang, G.; Liu, X.; Zhang, X.; Li, L.; Bian, R.; Cheng, K.; Zheng, J. A long-term hybrid poplar plantation on cropland reduces soil organic carbon mineralization and shifts microbial community abundance and composition. Appl. Soil Ecol. 2017, 111, 94–104. [Google Scholar] [CrossRef]
- Shen, Y.; Nie, J.; Li, Z.; Li, H.; Wu, Y.; Dong, Y.; Zhang, J. Differentiated surface fungal communities at point of harvest on apple fruits from rural and peri-urban orchards. Scientific reports 2018, 8, 2165. [Google Scholar] [CrossRef] [Green Version]
- Ecker, D.J.; Sampath, R.; Willett, P.; Wyatt, J.R.; Samant, V.; Massire, C.; Hall, T.A.; Hari, K.; McNeil, J.A.; Büchen-Osmond, C. The Microbial Rosetta Stone Database: A compilation of global and emerging infectious microorganisms and bioterrorist threat agents. BMC Microbiol. 2005, 5, 19. [Google Scholar] [CrossRef]
- Li, X.-G.; Ding, C.-F.; Zhang, T.-L.; Wang, X.-X. Fungal pathogen accumulation at the expense of plant-beneficial fungi as a consequence of consecutive peanut monoculturing. Soil Biol. Biochem. 2014, 72, 11–18. [Google Scholar] [CrossRef]
- Zhang, N.; Castlebury, L.A.; Miller, A.N.; Huhndorf, S.M.; Schoch, C.L.; Seifert, K.A.; Rossman, A.Y.; Rogers, J.D.; Kohlmeyer, J.; Volkmann-Kohlmeyer, B. An overview of the systematics of the Sordariomycetes based on a four-gene phylogeny. Mycologia 2006, 98, 1076–1087. [Google Scholar] [CrossRef]
- Soytong, K.; Kanokmedhakul, S.; Kukongviriyapa, V.; Isobe, M. Application of Chaetomium species (Ketomium) as a new broad spectrum biological fungicide for plant disease control. Fungal Divers 2001, 7, 1–15. [Google Scholar]
- Pandey, A. Studies on fungal diseases of eggplant in relation to statistical analysis and making of a disease calendar. Recent Res. Sci. Technol. 2010, 2. [Google Scholar]
- Xiong, W.; Zhao, Q.; Xue, C.; Xun, W.; Zhao, J.; Wu, H.; Li, R.; Shen, Q. Comparison of fungal community in black pepper-vanilla and vanilla monoculture systems associated with vanilla Fusarium wilt disease. Front. Microbiol. 2016, 7, 117. [Google Scholar] [CrossRef]
- Barkai-Golan, R. Postharvest Diseases of Fruits and Vegetables: Development and Control; Elsevier: Amsterdam, The Netherlands, 2001. [Google Scholar]
- Khan, A.L.; Al-Harrasi, A.; Al-Rawahi, A.; Al-Farsi, Z.; Al-Mamari, A.; Waqas, M.; Asaf, S.; Elyassi, A.; Mabood, F.; Shin, J.-H. Endophytic fungi from Frankincense tree improves host growth and produces extracellular enzymes and indole acetic acid. PloS ONE 2016, 11, e0158207. [Google Scholar] [CrossRef]
- Yao, Y.; Xue, Z.; Hong, C.; Zhu, F.; Chen, X.; Wang, W.; Cai, Z.; Huang, N.; Yang, X. Efficiency of different solarization-based ecological soil treatments on the control of Fusarium wilt and their impacts on the soil microbial community. Appl. Soil Ecol. 2016, 108, 341–351. [Google Scholar] [CrossRef]
- Nakasaki, K.; Saito, M.; Suzuki, N. Coprinellus curtus (Hitoyo-take) prevents diseases of vegetables caused by pathogenic fungi. FEMS Microbiol. Lett. 2007, 275, 286–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rai, B.; Upadhyay, R. Competitive saprophytic colonization of pigeon-pea substrate by Fusarium udum in relation to environmental factors, chemical treatments and microbial antagonism. Soil Biol. Biochem. 1983, 15, 187–191. [Google Scholar] [CrossRef]
- Kawaradani, M.; Taguchi, K.; Okada, K.; Hirooka, Y.; Sato, T. Seedling rot of garland chrysanthemum caused by Gibellulopsis chrysanthemi and ecological characters of the causal fungus. J. Gen. Plant Pathol. 2013, 79, 346–349. [Google Scholar] [CrossRef]
- Vesper, S.; Turner, J.; Phillips, D. Incidence of Verticillium nigrescens in soybeans. Phytopathology 1983, 73, 1338–1340. [Google Scholar] [CrossRef]
- Hamayun, M.; Khan, S.A.; Iqbal, I.; Na, C.-I.; Khan, A.L.; Hwang, Y.-H.; Lee, B.-H.; Lee, I.-J. Chrysosporium pseudomerdarium produces gibberellins and promotes plant growth. J. Microbiol. 2009, 47, 425–430. [Google Scholar] [CrossRef]
- van Elsas, J.D.; Chiurazzi, M.; Mallon, C.A.; Elhottovā, D.; Krištůfek, V.; Salles, J.F. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc. Natl. Acad. Sci. USA 2012, 109, 1159–1164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, A.; Ghani, M.I.; Ding, H.; Fan, Y.; Cheng, Z.; Iqbal, M. Co-Amended Synergistic Interactions between Arbuscular Mycorrhizal Fungi and the Organic Substrate-Induced Cucumber Yield and Fruit Quality Associated with the Regulation of the AM-Fungal Community Structure under Anthropogenic Cultivated Soil. Int. J. Mol. Sci. 2019, 20, 1539. [Google Scholar] [CrossRef]
- Wang, S.; Tian, H.; Liu, J.; Pan, S. Pattern and change of soil organic carbon storage in China: 1960s–1980s. Tellus B Chem. Phys. Meteorol. 2003, 55, 416–427. [Google Scholar]
- Parkinson, J.; Allen, S. A wet oxidation procedure suitable for the determination of nitrogen and mineral nutrients in biological material. Commun. Soil Sci. Plant Anal. 1975, 6, 1–11. [Google Scholar] [CrossRef]
- Olsen, S.R. Estimation of Available Phosphorus in Soils by Extraction with Sodium Bicarbonate; United States Department Of Agriculture: Washington, WA, USA, 1954.
- Knudsen, D.; Peterson, G.; Pratt, P. Lithium, sodium, and potassium. In Methods Soil Analysis Part 2; American Society of Agronomy: Madison, WI, USA, 1982; pp. 225–246. [Google Scholar]
- Guan, S.; Zhang, D.; Zhang, Z. Soil enzyme and its research methods. Agric. Beijing 1986, 1986, 274–297. [Google Scholar]
- Tabatabai, M. Enzymes. In Methods of Soil Analysis. Part 2. Microbiological and Biochemical Properties; Weaver, R.W., Augle, S., Bottomly, P.J., Berdicek, Q., Smith, Q., Tabatabai, A., Wollum, A., Eds.; Soil Science Society of America: Madison, WI, USA, 1994; pp. 775–833. [Google Scholar]














| Treatment | pH | EC (µs⋅cm−1) | Organic Carbon (g⋅kg−1) | Organic Matter (g⋅kg−1) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2016 | 2017 | Means | 2016 | 2017 | Means | 2016 | 2017 | Means | 2016 | 2017 | Means | |
| RGS0 | 7.89 | 7.91 | 7.90a | 164.24 | 162.52 | 163.41b | 15.21 | 16.09 | 15.66c | 26.22 | 27.76 | 26.99c |
| RGS1 | 7.85 | 7.80 | 7.82b | 228.50 | 230.67 | 229.58a | 19.27 | 20.36 | 19.18b | 33.21 | 35.10 | 34.16b |
| RGS2 | 7.78 | 7.76 | 7.77bc | 227.00 | 235.83 | 231.42a | 21.46 | 23.02 | 22.24a | 37.00 | 39.70 | 38.35a |
| RGS3 | 7.75 | 7.70 | 7.72c | 234.00 | 238.67 | 236.33a | 23.06 | 24.17 | 23.62a | 39.76 | 41.68 | 40.72a |
| Year means | 7.82 | 7.79 | 213.43 | 216.94 | 19.75 | 20.916 | 34.04 | 36.06 | ||||
| LSD-test | Treatment | Year | Interaction | Treatment | Year | Interaction | Treatment | Year | Interaction | Treatment | Year | Interaction |
| *** | NS | NS | *** | NS | NS | *** | NS | NS | *** | NS | NS | |
| Treatment | Available N (mg⋅kg−1) | Available P (mg⋅kg−1) | Available K (mg⋅kg−1) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 2016 | 2017 | Means | 2016 | 2017 | Means | 2016 | 2017 | Means | |
| RGS0 | 70.79 | 72.20 | 71.50b | 194.44 | 203.11 | 198.78c | 226.33 | 236.67 | 231.50b |
| RGS1 | 75.13 | 78.13 | 76.63b | 198.55 | 204.78 | 201.66c | 232.17 | 248.33 | 240.25b |
| RGS2 | 119.10 | 125.00 | 122.05a | 214.55 | 227.85 | 221.20b | 377.83 | 385.50 | 381.67a |
| RGS3 | 120.24 | 126.94 | 123.59a | 221.85 | 232.15 | 227.00a | 396.83 | 403.83 | 400.33a |
| Year Means | 96.31a | 100.57a | 207.35 | 216.97 | 308.29a | 318.58a | |||
| LSD-test | Treatment | Year | Interaction | Treatment | Year | Interaction | Treatment | Year | Interaction |
| *** | NS | NS | *** | NS | NS | *** | NS | NS | |
| Treatment | Soil Invertase Activity (Glucose mg g−1) | Soil Urease Activity (NH3-N mg.g−1) | Soil Dehydrogenase (mg TPF kg−1 soil h−1) | Soil Phosphatase (P2O5 mg.100g−1) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2016 | 2017 | Means | 2016 | 2017 | Means | 2016 | 2017 | Means | 2016 | 2017 | Means | |
| RGS0 | 88.77 | 91.75 | 90.26d | 1.67f | 3.19c | 2.43 | 15.61 | 16.21 | 15.91c | 5.80e | 6.12de | 5.96d |
| RGS1 | 101.85 | 110.30 | 106.07c | 2.10e | 4.69b | 2.40 | 19.26 | 19.74 | 19.50b | 6.77cd | 7.23c | 7.00c |
| RGS2 | 116.61 | 118.00 | 117.30b | 2.79d | 4.88ab | 3.83 | 29.20 | 31.52 | 30.27a | 8.52b | 9.26a | 8.89a |
| RGS3 | 129.31 | 134.88 | 132.10a | 3.03c | 5.08a | 4.06 | 24.59 | 25.78 | 25.18b | 8.01b | 8.66ab | 8.34b |
| Year means | 109.13 | 113.73 | 2.40 | 4.46 | 22.163 | 23.27 | 7.28 | 7.81 | ||||
| LSD-test | Treatment | Year | Interaction | Treatment | Year | Interaction | Treatment | Year | Interaction | Treatment | Year | Interaction |
| *** | NS | NS | *** | *** | *** | *** | NS | NS | *** | * | NS | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghani, M.I.; Ali, A.; Atif, M.J.; Ali, M.; Amin, B.; Anees, M.; Khurshid, H.; Cheng, Z. Changes in the Soil Microbiome in Eggplant Monoculture Revealed by High-Throughput Illumina MiSeq Sequencing as Influenced by Raw Garlic Stalk Amendment. Int. J. Mol. Sci. 2019, 20, 2125. https://doi.org/10.3390/ijms20092125
Ghani MI, Ali A, Atif MJ, Ali M, Amin B, Anees M, Khurshid H, Cheng Z. Changes in the Soil Microbiome in Eggplant Monoculture Revealed by High-Throughput Illumina MiSeq Sequencing as Influenced by Raw Garlic Stalk Amendment. International Journal of Molecular Sciences. 2019; 20(9):2125. https://doi.org/10.3390/ijms20092125
Chicago/Turabian StyleGhani, Muhammad Imran, Ahmad Ali, Muhammad Jawaad Atif, Muhammad Ali, Bakht Amin, Muhammad Anees, Haris Khurshid, and Zhihui Cheng. 2019. "Changes in the Soil Microbiome in Eggplant Monoculture Revealed by High-Throughput Illumina MiSeq Sequencing as Influenced by Raw Garlic Stalk Amendment" International Journal of Molecular Sciences 20, no. 9: 2125. https://doi.org/10.3390/ijms20092125
APA StyleGhani, M. I., Ali, A., Atif, M. J., Ali, M., Amin, B., Anees, M., Khurshid, H., & Cheng, Z. (2019). Changes in the Soil Microbiome in Eggplant Monoculture Revealed by High-Throughput Illumina MiSeq Sequencing as Influenced by Raw Garlic Stalk Amendment. International Journal of Molecular Sciences, 20(9), 2125. https://doi.org/10.3390/ijms20092125