More P450s Are Involved in Secondary Metabolite Biosynthesis in Streptomyces Compared to Bacillus, Cyanobacteria, and Mycobacterium
Abstract
:1. Introduction
2. Results and Discussion
2.1. Streptomyces Species Have Large Number of P450s
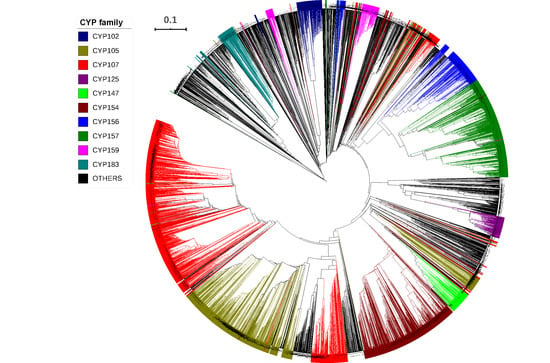
2.2. CYP107 Family Was Found to Be Dominant and Conserved in 203 Streptomyces Species
2.3. Numerous P450s Involved in Secondary Metabolite Production in Streptomyces Compared to Other Bacterial Species
| P450 | Species | Function | References |
|---|---|---|---|
| CYP158A1 | Streptomyces coelicolor A3(2) | Flaviolin biosynthesis | [38] |
| CYP1048A1 | Streptomyces scabiei | Thaxtomin (phytotoxin) biosynthesis | [39] |
| CYP105A1 | Streptomyces griseolus | Diterpenoids synthesis | [40] |
| CYP105A3 (P450sca-2) | Streptomyces carbophilus | Pravastatin synthesis | [41] |
| CYP105B28(GfsF) * | Streptomyces graminofaciens | Macrolide antibiotic synthesis | [42,43] |
| CYP105D6 | Streptomyces avermitilis | Filipin biosynthesis | [44] |
| CYP105D7 | Streptomyces avermitilis | Filipin biosynthesis | [45] |
| CYP105D8 | Streptomyces tubercidicus strain I-1529 | Avermectin oxidation | [32,46] |
| CYP105D9 | Streptomyces sp. JP95 | Griseorhodin biosynthesis | [32,47] |
| CYP105F2 | Streptomyces peucetius | Oleandomycin biosynthesis | [48,49] |
| CYP105H1 | Streptomyces noursei ATCC 11455 | Nystatin biosynthesis | [32] |
| CYP105H3 | Streptomyces natalensis | Pimaricin biosynthesis | [32,50] |
| CYP105H4 (AmphN) ! | Streptomyces nodosus | Amphotericin biosynthesis | [51,52] |
| CYP105H5 | Streptomyces griseus | Candidicin biosynthesis | [32] |
| CYP105K1 | Streptomyces tendae strain Tue901 | Nikkomycin biosynthesis | [32,53] |
| CYP105K2 | Streptomyces ansochromogenes | Nikkomycin biosynthesis | [32] |
| CYP105L1 (TylH1,orf7) ! | Streptomyces fradiae | Tylosin biosynthesis | [54,55] |
| CYP105L4(ChmH1) * | Streptomyces bikiniensis | Chalcomycin biosynthesis | [56] |
| CYP105M1 (orf10) ! | Streptomyces clavuligerus | Clavulanic acid antibiotic biosynthesis | [57] |
| CYP105N1 | Streptomyces coelicolor A3(2) | Coelibactin siderophore biosynthesis | [58,59] |
| CYP105P1 | Streptomyces avermitilis | Filipin biosynthesis | [44] |
| CYP105U1 | Streptomyces hygroscopicus | Geldanamycin biosynthesis | [60] |
| CYP105V1 | Streptomyces sp. HK803 | Phoslactomycin biosynthesis | [32,61] |
| CYP105AA1 | Streptomyces tubercidicus strain R922 | Avermectin oxidation | [32,46] |
| CYP105AA2 | Streptomyces tubercidicus strain I-1529 | Avermectin oxidation | [32,46] |
| CYP107A1 | Streptomyces peucetius | Dealkylation of 7-ethoxycoumarin | [62] |
| CYP107A1 | Saccharopolyspora erythraea | Erythromycin biosynthesis | [63,64] |
| CYP107B (HmtN) ! | Streptomyces himastatinicus ATCC 53653 | Himastatin biosynthesis | [65] |
| CYP107B (HmtN) | Streptomyces himastatinicus | Himastatin biosynthesis | [66] |
| CYP107C1 | Streptomyces thermotolerans | Carbomycin biosynthesis | [67] |
| CYP107E40(chmPII) * | Streptomyces bikiniensis | Chalcomycin biosynthesis | [56] |
| CYP107EE2(chmPI) * | Streptomyces bikiniensis | Chalcomycin biosynthesis | [56] |
| CYP107FH5(TamI) * | Streptomyces sp. 307-9 | Tirandamycin biosynthesis | [68,69] |
| CYP107G1 | Streptomyces rapamycinicus | Rapamycin biosynthesis | [70,71] |
| CYP107G1 (rapN) ! | Streptomyces hygroscopicus | Rapamycin biosynthesis | [71,72] |
| CYP107L1 | Streptomyces venezuelae | Macrolide antibioitics biosynthesis | [73] |
| CYP107L59(FosK) * | Streptomyces pulveraceus | Fostriecin biosynthesis | [74] |
| CYP107MD3(FosG) * | Streptomyces pulveraceus | Fostriecin biosynthesis | [74] |
| CYP107W1 | Streptomyces avermitilis | Oligomycin A biosynthesis | [75,76] |
| CYP112A2 | Streptomyces rapamycinicus | Rapamycin biosynthesis | [70,71] |
| CYP113A1 | Saccharopolyspora erythraea | Erythromycin biosynthesis | [63,64] |
| CYP113B1 (TylI) ! | Streptomyces fradiae | Tylosin biosynthesis | [54,55] |
| CYP113D3(HmtT) * | Streptomyces himastatinicus ATCC 53653 | Himastatin biosynthesis | [65] |
| CYP113D3 (HmtT) * | Streptomyces himastatinicus | Himastatin biosynthesis | [66] |
| CYP113HI (HmtS) * | Streptomyces himastatinicus | Himastatin biosynthesis | [66] |
| CYP122A2 (rapJ) ! | Streptomyces hygroscopicus | Rapamycin biosynthesis | [70,71] |
| CYP122A3 | Streptomyces hygroscopicus | Rapamycin biosynthesis | [70,71] |
| CYP122A4 (FkbD) ! | Streptomyces tsukubaensis | FK506 (immunosuppressant) polyketide biosynthesis | [77] |
| CYP129A2 | Streptomyces peucetius | Doxorubicin biosynthesis | [78,79] |
| CYP129A2 (dox A) ! | Streptomyces sp. strain C5 | Doxorubicin biosynthesis | [80,81] |
| CYP131A2 (dnrQ) ! | Streptomyces sp. strain C5 | Doxorubicin biosynthesis | [80,81] |
| CYP140M1(TtnI) * | Streptomyces griseochromogenes | Tautomycetin biosynthesis | [82] |
| CYP151A (AurH) ! | Streptomyces thioluteus | Aureothin biosynthesis | [83] |
| CYP154A1 | Streptomyces coelicolor A3(2) | Polyketide synthesis and cyclization of a cellular dipentaenone | [84,85] |
| CYP154B1 | Streptomyces fradiae | Tylosin biosynthesis | [54,55] |
| CYP154C1 | Streptomyces coelicolor A3(2) | Macrolide biosynthesis | [86] |
| CYP158A2 | Streptomyces coelicolor A3(2) | Flaviolin biosynthesis | [87] |
| CYP161A2 (PimD) ! | Streptomyces natalensis | Pimaricin biosynthesis | [88] |
| CYP161A3 (AmphL) ! | Streptomyces nodosus | Amphotericin biosynthesis | [51] |
| CYP162A1 | Streptomyces tendae | Nikkomycin biosynthesis | [53,89] |
| CYP163A1 (NovI) ! | Streptomyces spheroids | Novobiocin biosynthesis | [90] |
| CYP163B3 (P450 Sky) ! | Streptomyces sp. Acta 2897 | Skyllamycin biosynthesis | [91] |
| CYP170A1 | Streptomyces coelicolor A3(2) | Albaflavenone biosynthesis | [92] |
| CYP170A2 | Streptomyces avermitilis | Albaflavenone biosynthesis | [93] |
| CYP170B1 | Streptomyces albus | Albaflavenone biosynthesis | [94] |
| CYP171A1 | Streptomyces avermitilis | Avermectin biosynthesis | [95,96] |
| CYP183A1 | Streptomyces avermitilis | Pentalenolactone biosynthesis | [96,97] |
| CYP244A1 (StaN) ! | Streptomyces sp tp-a0274 | Rapamycin biosynthesis | [70,71] |
| CYP245A1 (StaP) ! | Streptomyces sp tp-a0274 | Rapamycin biosynthesis | [70,71] |
| CYP246A1 | Streptomyces scabiei | Thaxtomin (phytotoxin) biosynthesis | [98] |
| CYP248A1 | Streptomyces thioluteus | Aureothin biosynthesis | [83] |
3. Materials and Methods
3.1. Information on Streptomyces Species and Genome Database
3.2. Genome Data Mining and Identification of P450s
3.3. Allocating Family and Subfamily to P450s
3.4. Streptomyces P450 Phylogenetic Analysis
3.5. Streptomyces P450 Profile Heat-Maps
3.6. Identification of P450s That Are Part of Secondary Metabolite BGCs
3.7. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Urlacher, V.B.; Girhard, M. Cytochrome P450 Monooxygenases in Biotechnology and Synthetic Biology. Trends Biotechnol. 2019, 37, 882–897. [Google Scholar] [CrossRef]
- Poulos, T.L.; Finzel, B.C.; Howard, A.J. High-resolution crystal structure of cytochrome P450cam. J. Mol. Biol. 1987, 195, 687–700. [Google Scholar] [CrossRef]
- Garfinkel, D. Studies on pig liver microsomes. I. Enzymic and pigment composition of different microsomal fractions. Arch. Biochem. Biophys. 1958, 77, 493–509. [Google Scholar] [CrossRef]
- Klingenberg, M. Pigments of rat liver microsomes. Arch Biochem. Biophys. 1958, 75, 376–386. [Google Scholar] [CrossRef]
- Omura, T. Recollection of the early years of the research on cytochrome P450. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2011, 87, 617–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omura, T.; Sato, R. A new cytochrome in liver microsomes. J. Biol. Chem. 1962, 237, 1375–1376. [Google Scholar]
- Nelson, D.R. Cytochrome P450 diversity in the tree of life. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 141–154. [Google Scholar] [CrossRef]
- Lamb, D.C.; Follmer, A.H.; Goldstone, J.V.; Nelson, D.R.; Warillow, A.G.; Price, C.L.; True, M.Y.; Kelly, S.L.; Poulos, T.L.; Stegeman, J.J. On the occurrence of cytochrome P450 in viruses. Proc. Natl. Acad. Sci. USA 2019, 116, 12343–12352. [Google Scholar] [CrossRef] [Green Version]
- Lepesheva, G.I.; Hargrove, T.Y.; Kleshchenko, Y.; Nes, W.D.; Villalta, F.; Waterman, M.R. CYP51: A major drug target in the cytochrome P450 superfamily. Lipids 2008, 43, 1117–1125. [Google Scholar] [CrossRef] [Green Version]
- Kelly, S.L.; Kelly, D.E. Microbial cytochromes P450: Biodiversity and biotechnology. Where do cytochromes P450 come from, what do they do and what can they do for us? Philos. Trans. R. Soc. London Ser. B Biol. Sci. 2013, 368, 20120476. [Google Scholar] [CrossRef] [Green Version]
- Jawallapersand, P.; Mashele, S.S.; Kovacic, L.; Stojan, J.; Komel, R.; Pakala, S.B.; Krasevec, N.; Syed, K. Cytochrome P450 monooxygenase CYP53 family in fungi: Comparative structural and evolutionary analysis and its role as a common alternative anti-fungal drug target. PLoS ONE 2014, 9, e107209. [Google Scholar] [CrossRef] [Green Version]
- Ziniel, P.D.; Karumudi, B.; Barnard, A.H.; Fisher, E.M.; Thatcher, G.R.; Podust, L.M.; Williams, D.L. The Schistosoma mansoni Cytochrome P450 (CYP3050A1) Is Essential for Worm Survival and Egg Development. PLoS Negl. Trop. Dis. 2015, 9, e0004279. [Google Scholar] [CrossRef]
- Ortiz de Montellano, P.R. Potential drug targets in the Mycobacterium tuberculosis cytochrome P450 system. J. Inorg. Biochem. 2018, 180, 235–245. [Google Scholar] [CrossRef]
- Podust, L.M.; Sherman, D.H. Diversity of P450 enzymes in the biosynthesis of natural products. Nat. Prod. Rep. 2012, 29, 1251–1266. [Google Scholar] [CrossRef] [Green Version]
- Greule, A.; Stok, J.E.; De Voss, J.J.; Cryle, M.J. Unrivalled diversity: The many roles and reactions of bacterial cytochromes P450 in secondary metabolism. Nat. Prod. Rep. 2018, 35, 757–791. [Google Scholar] [CrossRef] [Green Version]
- Rudolf, J.D.; Chang, C.Y.; Ma, M.; Shen, B. Cytochromes P450 for natural product biosynthesis in Streptomyces: Sequence, structure, and function. Nat. Prod. Rep. 2017, 34, 1141–1172. [Google Scholar] [CrossRef]
- Nelson, D.R.; Kamataki, T.; Waxman, D.J.; Guengerich, F.P.; Estabrook, R.W.; Feyereisen, R.; Gonzalez, F.J.; Coon, M.J.; Gunsalus, I.C.; Gotoh, O.; et al. The P450 superfamily: Update on new sequences, gene mapping, accession numbers, early trivial names of enzymes, and nomenclature. DNA Cell Biol. 1993, 12, 1–51. [Google Scholar] [CrossRef]
- Nelson, D.R. Cytochrome P450 nomenclature. Methods Mol. Biol. (Clifton NJ) 1998, 107, 15–24. [Google Scholar] [CrossRef]
- Nelson, D.R. Cytochrome P450 nomenclature, 2004. Methods Mol. Biol. (Clifton NJ) 2006, 320, 1–10. [Google Scholar] [CrossRef]
- Senate, L.M.; Tjatji, M.P.; Pillay, K.; Chen, W.; Zondo, N.M.; Syed, P.R.; Mnguni, F.C.; Chiliza, Z.E.; Bamal, H.D.; Karpoormath, R. Similarities, variations, and evolution of cytochrome P450s in Streptomyces versus Mycobacterium. Sci. Rep. 2019, 9, 3962. [Google Scholar] [CrossRef] [Green Version]
- Parvez, M.; Qhanya, L.B.; Mthakathi, N.T.; Kgosiemang, I.K.; Bamal, H.D.; Pagadala, N.S.; Xie, T.; Yang, H.; Chen, H.; Theron, C.W.; et al. Molecular evolutionary dynamics of cytochrome P450 monooxygenases across kingdoms: Special focus on mycobacterial P450s. Sci. Rep. 2016, 6, 33099. [Google Scholar] [CrossRef] [Green Version]
- Mthethwa, B.; Chen, W.; Ngwenya, M.; Kappo, A.; Syed, P.; Karpoormath, R.; Yu, J.-H.; Nelson, D.; Syed, K. Comparative analyses of cytochrome P450s and those associated with secondary metabolism in Bacillus species. Int. J. Mol. Sci. 2018, 19, 3623. [Google Scholar] [CrossRef] [Green Version]
- Khumalo, M.J.N.; Padayachee, T.; Chen, W.; Yu, J.-H.; Nelson, D.; Syed, K. Comprehensive analyses of cytochrome P450 monoxygenases and secondary metabolite biosynthetic gene clusters in Cyanobacteria. Int. J. Mol. Sci. 2020, 21, 656. [Google Scholar] [CrossRef] [Green Version]
- Syed, K.; Shale, K.; Pagadala, N.S.; Tuszynski, J. Systematic identification and evolutionary analysis of catalytically versatile cytochrome P450 monooxygenase families enriched in model basidiomycete fungi. PLoS ONE 2014, 9, e86683. [Google Scholar] [CrossRef]
- Kgosiemang, I.K.R.; Syed, K.; Mashele, S.S. Comparative genomics and evolutionary analysis of cytochrome P450 monooxygenases in fungal subphylum Saccharomycotina. J. Pure Appl. Microbiol. 2014, 8, 12. [Google Scholar]
- Sello, M.M.; Jafta, N.; Nelson, D.R.; Chen, W.; Yu, J.H.; Parvez, M.; Kgosiemang, I.K.; Monyaki, R.; Raselemane, S.C.; Qhanya, L.B.; et al. Diversity and evolution of cytochrome P450 monooxygenases in Oomycetes. Sci. Rep. 2015, 5, 11572. [Google Scholar] [CrossRef] [Green Version]
- Akapo, O.O.; Padayachee, T.; Chen, W.; Kappo, A.P.; Yu, J.H.; Nelson, D.R.; Syed, K. Distribution and Diversity of Cytochrome P450 Monooxygenases in the Fungal Class Tremellomycetes. Int. J. Mol. Sci. 2019, 20, 2889. [Google Scholar] [CrossRef] [Green Version]
- de Lima Procópio, R.E.; da Silva, I.R.; Martins, M.K.; de Azevedo, J.L.; de Araújo, J.M. Antibiotics produced by Streptomyces. Braz. J. Infect. Dis. 2012, 16, 466–471. [Google Scholar] [CrossRef] [Green Version]
- Hwang, K.-S.; Kim, H.U.; Charusanti, P.; Palsson, B.Ø.; Lee, S.Y. Systems biology and biotechnology of Streptomyces species for the production of secondary metabolites. Biotechnol. Adv. 2014, 32, 255–268. [Google Scholar] [CrossRef]
- Harir, M.; Bendif, H.; Bellahcene, M.; Fortas, Z.; Pogni, R. Streptomyces Secondary Metabolites. In Basic Biology and Applications of Actinobacteria; IntechOpen: London, UK, 2018; pp. 99–122. [Google Scholar] [CrossRef] [Green Version]
- Cho, M.-A.; Han, S.; Lim, Y.-R.; Kim, V.; Kim, H.; Kim, D. Streptomyces Cytochrome P450 Enzymes and Their Roles in the Biosynthesis of Macrolide Therapeutic Agents. Biomol. Ther. 2019, 27, 127. [Google Scholar] [CrossRef]
- Moody, S.C.; Loveridge, E.J. CYP105-diverse structures, functions and roles in an intriguing family of enzymes in Streptomyces. J. Appl. Microbiol. 2014, 117, 1549–1563. [Google Scholar] [CrossRef] [Green Version]
- Lamb, D.C.; Waterman, M.R.; Zhao, B. Streptomyces cytochromes P450: Applications in drug metabolism. Expert Opin. Drug Metab. Toxicol. 2013, 9, 1279–1294. [Google Scholar] [CrossRef]
- Feyereisen, R. Insect CYP genes and P450 enzymes. In Insect Molecular Biology and Biochemistry; Elsevier: Amsterdam, The Netherlands, 2012; pp. 236–316. [Google Scholar]
- Qhanya, L.B.; Matowane, G.; Chen, W.; Sun, Y.; Letsimo, E.M.; Parvez, M.; Yu, J.H.; Mashele, S.S.; Syed, K. Genome-wide annotation and comparative analysis of cytochrome P450 monooxygenases in Basidiomycete biotrophic plant pathogens. PLoS ONE 2015, 10, e0142100. [Google Scholar] [CrossRef] [Green Version]
- Ngwenya, M.L.; Chen, W.; Basson, A.K.; Shandu, J.S.; Yu, J.H.; Nelson, D.R.; Syed, K. Blooming of unusual cytochrome P450s by tandem duplication in the pathogenic fungus Conidiobolus coronatus. Int. J. Mol. Sci. 2018, 19, 1711. [Google Scholar] [CrossRef] [Green Version]
- Tran, P.N.; Yen, M.R.; Chiang, C.Y.; Lin, H.C.; Chen, P.Y. Detecting and prioritizing biosynthetic gene clusters for bioactive compounds in bacteria and fungi. Appl. Microbiol. Biotechnol. 2019, 103, 3277–3287. [Google Scholar] [CrossRef] [Green Version]
- Zhao, B.; Lamb, D.C.; Lei, L.; Kelly, S.L.; Yuan, H.; Hachey, D.L.; Waterman, M.R. Different binding modes of two flaviolin substrate molecules in cytochrome P450 158A1 (CYP158A1) compared to CYP158A2. Biochemistry 2007, 46, 8725–8733. [Google Scholar] [CrossRef]
- Yu, F.; Li, M.; Xu, C.; Wang, Z.; Zhou, H.; Yang, M.; Chen, Y.; Tang, L.; He, J. Structural insights into the mechanism for recognizing substrate of the cytochrome P450 enzyme TxtE. PLoS ONE 2013, 8, e81526. [Google Scholar] [CrossRef] [Green Version]
- Janocha, S.; Zapp, J.; Hutter, M.; Kleser, M.; Bohlmann, J.; Bernhardt, R. Resin acid conversion with CYP105A1: An enzyme with potential for the production of pharmaceutically relevant diterpenoids. Chembiochem 2013, 14, 467–473. [Google Scholar] [CrossRef]
- Watanabe, I.; Nara, F.; Serizawa, N. Cloning, characterization and expression of the gene encoding cytochrome P-450sca-in2 from Streptomyces carbophilus involved in production of pravastatin, a specific HMG-CoA reductase inhibitor. Gene 1995, 163, 81–85. [Google Scholar] [CrossRef]
- Kudo, F.; Motegi, A.; Mizoue, K.; Eguchi, T. Cloning and characterization of the biosynthetic gene cluster of 16-membered macrolide antibiotic FD-891: Involvement of a dual functional cytochrome P450 monooxygenase catalyzing epoxidation and hydroxylation. Chembiochem 2010, 11, 1574–1582. [Google Scholar] [CrossRef]
- Kataoka, T.; Yamada, A.; Bando, M.; Honma, T.; Mizoue, K.; Nagai, K. FD-891, a structural analogue of concanamycin A that does not affect vacuolar acidification or perforin activity, yet potently prevents cytotoxic T lymphocyte-mediated cytotoxicity through the blockage of conjugate formation. Immunology 2000, 100, 170–177. [Google Scholar] [CrossRef]
- Xu, L.H.; Fushinobu, S.; Takamatsu, S.; Wakagi, T.; Ikeda, H.; Shoun, H. Regio- and stereospecificity of filipin hydroxylation sites revealed by crystal structures of cytochrome P450 105P1 and 105D6 from Streptomyces avermitilis. J. Biol. Chem. 2010, 285, 16844–16853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takamatsu, S.; Xu, L.H.; Fushinobu, S.; Shoun, H.; Komatsu, M.; Cane, D.E.; Ikeda, H. Pentalenic acid is a shunt metabolite in the biosynthesis of the pentalenolactone family of metabolites: Hydroxylation of 1-deoxypentalenic acid mediated by CYP105D7 (SAV_7469) of Streptomyces avermitilis. J. Antibiot. 2011, 64, 65–71. [Google Scholar] [CrossRef] [Green Version]
- Jungmann, V.; Molnar, I.; Hammer, P.E.; Hill, D.S.; Zirkle, R.; Buckel, T.G.; Buckel, D.; Ligon, J.M.; Pachlatko, J.P. Biocatalytic conversion of avermectin to 4″-oxo-avermectin: Characterization of biocatalytically active bacterial strains and of cytochrome p450 monooxygenase enzymes and their genes. Appl. Environ. Microbiol. 2005, 71, 6968–6976. [Google Scholar] [CrossRef] [Green Version]
- Yunt, Z.; Reinhardt, K.; Li, A.; Engeser, M.; Dahse, H.M.; Gutschow, M.; Bruhn, T.; Bringmann, G.; Piel, J. Cleavage of four carbon-carbon bonds during biosynthesis of the griseorhodin a spiroketal pharmacophore. J. Am. Chem. Soc. 2009, 131, 2297–2305. [Google Scholar] [CrossRef]
- Rodriguez, A.M.; Olano, C.; Mendez, C.; Hutchinson, C.R.; Salas, J.A. A cytochrome P450-like gene possibly involved in oleandomycin biosynthesis by Streptomyces antibioticus. FEMS Microbiol. Lett. 1995, 127, 117–120. [Google Scholar] [CrossRef]
- Shrestha, P.; Oh, T.J.; Liou, K.; Sohng, J.K. Cytochrome P450 (CYP105F2) from Streptomyces peucetius and its activity with oleandomycin. Appl. Microbiol. Biotechnol. 2008, 79, 555–562. [Google Scholar] [CrossRef]
- Aparicio, J.F.; Fouces, R.; Mendes, M.V.; Olivera, N.; Martin, J.F. A complex multienzyme system encoded by five polyketide synthase genes is involved in the biosynthesis of the 26-membered polyene macrolide pimaricin in Streptomyces natalensis. Chem. Biol. 2000, 7, 895–905. [Google Scholar] [CrossRef] [Green Version]
- Caffrey, P.; Lynch, S.; Flood, E.; Finnan, S.; Oliynyk, M. Amphotericin biosynthesis in Streptomyces nodosus: Deductions from analysis of polyketide synthase and late genes. Chem. Biol. 2001, 8, 713–723. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, P.K.; Agarwal, P.; Reddy, M.K.; Sopory, S.K. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006, 25, 1263–1274. [Google Scholar] [CrossRef]
- Lauer, B.; Russwurm, R.; Schwarz, W.; Kalmanczhelyi, A.; Bruntner, C.; Rosemeier, A.; Bormann, C. Molecular characterization of co-transcribed genes from Streptomyces tendae Tu901 involved in the biosynthesis of the peptidyl moiety and assembly of the peptidyl nucleoside antibiotic nikkomycin. Mol. Gen. Genet. MGG 2001, 264, 662–673. [Google Scholar] [CrossRef]
- Merson-Davies, L.A.; Cundliffe, E. Analysis of five tylosin biosynthetic genes from the tyllBA region of the Streptomyces fradiae genome. Mol. Microbiol. 1994, 13, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Fouces, R.; Mellado, E.; Diez, B.; Barredo, J.L. The tylosin biosynthetic cluster from Streptomyces fradiae: Genetic organization of the left region. Microbiology 1999, 145 Pt 4, 855–868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, S.L.; Hu, Z.; Schirmer, A.; Reid, R.; Revill, W.P.; Reeves, C.D.; Petrakovsky, O.V.; Dong, S.D.; Katz, L. Chalcomycin biosynthesis gene cluster from Streptomyces bikiniensis: Novel features of an unusual ketolide produced through expression of the chm polyketide synthase in Streptomyces fradiae. Antimicrob. Agents Chemother. 2004, 48, 4703–4712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reading, C.; Cole, M. Clavulanic acid: A beta-lactamase-inhibiting beta-lactam from Streptomyces clavuligerus. Antimicrob. Agents Chemother. 1977, 11, 852–857. [Google Scholar] [CrossRef] [Green Version]
- Lim, Y.R.; Hong, M.K.; Kim, J.K.; Doan, T.T.; Kim, D.H.; Yun, C.H.; Chun, Y.J.; Kang, L.W.; Kim, D. Crystal structure of cytochrome P450 CYP105N1 from Streptomyces coelicolor, an oxidase in the coelibactin siderophore biosynthetic pathway. Arch Biochem. Biophys. 2012, 528, 111–117. [Google Scholar] [CrossRef]
- Zhao, B.; Moody, S.C.; Hider, R.C.; Lei, L.; Kelly, S.L.; Waterman, M.R.; Lamb, D.C. Structural analysis of cytochrome P450 105N1 involved in the biosynthesis of the zincophore, coelibactin. Int. J. Mol. Sci. 2012, 13, 8500–8513. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Ni, S.; Jia, C.; Wang, H.; Sun, G.; Wu, L.; Gan, M.; Shan, G.; He, W.; Lin, L.; et al. Identification of 4,5-dihydro-4-hydroxygeldanamycins as shunt products of geldanamycin biosynthesis. J. Nat. Prod. 2012, 75, 1480–1484. [Google Scholar] [CrossRef]
- Palaniappan, N.; Kim, B.S.; Sekiyama, Y.; Osada, H.; Reynolds, K.A. Enhancement and selective production of phoslactomycin B, a protein phosphatase II a inhibitor, through identification and engineering of the corresponding biosynthetic gene cluster. J. Biol. Chem. 2003, 278, 35552–35557. [Google Scholar] [CrossRef] [Green Version]
- Niraula, N.P.; Kanth, B.K.; Sohng, J.K.; Oh, T.J. Hydrogen peroxide-mediated dealkylation of 7-ethoxycoumarin by cytochrome P450 (CYP107AJ1) from Streptomyces peucetius ATCC27952. Enzym. Microb. Technol. 2011, 48, 181–186. [Google Scholar] [CrossRef]
- Shafiee, A.; Hutchinson, C.R. Macrolide antibiotic biosynthesis: Isolation and properties of two forms of 6-deoxyerythronolide B hydroxylase from Saccharopolyspora erythraea (Streptomyces erythreus). Biochemistry 1987, 26, 6204–6210. [Google Scholar] [CrossRef]
- Stassi, D.; Donadio, S.; Staver, M.J.; Katz, L. Identification of a Saccharopolyspora erythraea gene required for the final hydroxylation step in erythromycin biosynthesis. J. Bacteriol. 1993, 175, 182–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Chen, J.; Wang, H.; Xie, Y.; Ju, J.; Yan, Y.; Zhang, H. Structural analysis of HmtT and HmtN involved in the tailoring steps of himastatin biosynthesis. FEBS Lett. 2013, 587, 1675–1680. [Google Scholar] [CrossRef]
- Ma, J.; Wang, Z.; Huang, H.; Luo, M.; Zuo, D.; Wang, B.; Sun, A.; Cheng, Y.Q.; Zhang, C.; Ju, J. Biosynthesis of himastatin: Assembly line and characterization of three cytochrome P450 enzymes involved in the post-tailoring oxidative steps. Angew. Chem. (Int. Ed. Engl.) 2011, 50, 7797–7802. [Google Scholar] [CrossRef]
- Arisawa, A.; Tsunekawa, H.; Okamura, K.; Okamoto, R. Nucleotide sequence analysis of the carbomycin biosynthetic genes including the 3-O-acyltransferase gene from Streptomyces thermotolerans. Biosci. Biotechnol. Biochem. 1995, 59, 582–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlson, J.C.; Fortman, J.L.; Anzai, Y.; Li, S.; Burr, D.A.; Sherman, D.H. Identification of the tirandamycin biosynthetic gene cluster from Streptomyces sp. 307-9. Chembiochem 2010, 11, 564–572. [Google Scholar] [CrossRef] [Green Version]
- Carlson, J.C.; Li, S.; Gunatilleke, S.S.; Anzai, Y.; Burr, D.A.; Podust, L.M.; Sherman, D.H. Tirandamycin biosynthesis is mediated by co-dependent oxidative enzymes. Nat. Chem. 2011, 3, 628–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aparicio, J.F.; Molnar, I.; Schwecke, T.; Konig, A.; Haydock, S.F.; Khaw, L.E.; Staunton, J.; Leadlay, P.F. Organization of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: Analysis of the enzymatic domains in the modular polyketide synthase. Gene 1996, 169, 9–16. [Google Scholar] [CrossRef]
- Molnar, I.; Aparicio, J.F.; Haydock, S.F.; Khaw, L.E.; Schwecke, T.; Konig, A.; Staunton, J.; Leadlay, P.F. Organisation of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: Analysis of genes flanking the polyketide synthase. Gene 1996, 169, 1–7. [Google Scholar] [CrossRef]
- Huang, S.; Bjornsti, M.-A.; Houghton, P.J. Rapamycins: Mechanisms of action and cellular resistance. Cancer Biol. Ther. 2003, 2, 222–232. [Google Scholar] [CrossRef] [Green Version]
- Sherman, D.H.; Li, S.; Yermalitskaya, L.V.; Kim, Y.; Smith, J.A.; Waterman, M.R.; Podust, L.M. The structural basis for substrate anchoring, active site selectivity, and product formation by P450 PikC from Streptomyces venezuelae. J. Biol. Chem. 2006, 281, 26289–26297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, R.; Liu, X.; Su, C.; Ma, C.; Qiu, R.; Tang, L. Elucidation of the biosynthetic gene cluster and the post-PKS modification mechanism for fostriecin in Streptomyces pulveraceus. Chem. Biol. 2013, 20, 45–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, S.; Pham, T.V.; Kim, J.H.; Lim, Y.R.; Park, H.G.; Cha, G.S.; Yun, C.H.; Chun, Y.J.; Kang, L.W.; Kim, D. Functional characterization of CYP107W1 from Streptomyces avermitilis and biosynthesis of macrolide oligomycin A. Arch Biochem. Biophys. 2015, 575, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Pham, T.V.; Kim, J.H.; Lim, Y.R.; Park, H.G.; Cha, G.S.; Yun, C.H.; Chun, Y.J.; Kang, L.W.; Kim, D. Structural Analysis of the Streptomyces avermitilis CYP107W1-Oligomycin A Complex and Role of the Tryptophan 178 Residue. Mol. Cells 2016, 39, 211–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, D.; Zhang, Q.; Zhang, Q.; Cen, P.; Xu, Z.; Liu, W. Improvement of FK506 production in Streptomyces tsukubaensis by genetic enhancement of the supply of unusual polyketide extender units via utilization of two distinct site-specific recombination systems. Appl. Environ. Microbiol. 2012, 78, 5093–5103. [Google Scholar] [CrossRef] [Green Version]
- Lomovskaya, N.; Otten, S.L.; Doi-Katayama, Y.; Fonstein, L.; Liu, X.C.; Takatsu, T.; Inventi-Solari, A.; Filippini, S.; Torti, F.; Colombo, A.L.; et al. Doxorubicin overproduction in Streptomyces peucetius: Cloning and characterization of the dnrU ketoreductase and dnrV genes and the doxA cytochrome P-450 hydroxylase gene. J. Bacteriol. 1999, 181, 305–318. [Google Scholar] [CrossRef] [Green Version]
- Madduri, K.; Hutchinson, C.R. Functional characterization and transcriptional analysis of a gene cluster governing early and late steps in daunorubicin biosynthesis in Streptomyces peucetius. J. Bacteriol. 1995, 177, 3879–3884. [Google Scholar] [CrossRef] [Green Version]
- Dickens, M.L.; Priestley, N.D.; Strohl, W.R. In vivo and in vitro bioconversion of epsilon-rhodomycinone glycoside to doxorubicin: Functions of DauP, DauK, and DoxA. J. Bacteriol. 1997, 179, 2641–2650. [Google Scholar] [CrossRef] [Green Version]
- Walczak, R.J.; Dickens, M.L.; Priestley, N.D.; Strohl, W.R. Purification, properties, and characterization of recombinant Streptomyces sp. strain C5 DoxA, a cytochrome P-450 catalyzing multiple steps in doxorubicin biosynthesis. J. Bacteriol. 1999, 181, 298–304. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Luo, Y.; Ju, J.; Rajski, S.R.; Osada, H.; Shen, B. Characterization of the tautomycetin biosynthetic gene cluster from Streptomyces griseochromogenes provides new insight into dialkylmaleic anhydride biosynthesis. J. Nat. Prod. 2009, 72, 450–459. [Google Scholar] [CrossRef] [Green Version]
- Zocher, G.; Richter, M.E.; Mueller, U.; Hertweck, C. Structural fine-tuning of a multifunctional cytochrome P450 monooxygenase. J. Am. Chem. Soc. 2011, 133, 2292–2302. [Google Scholar] [CrossRef] [PubMed]
- Podust, L.M.; Bach, H.; Kim, Y.; Lamb, D.C.; Arase, M.; Sherman, D.H.; Kelly, S.L.; Waterman, M.R. Comparison of the 1.85 A structure of CYP154A1 from Streptomyces coelicolor A3(2) with the closely related CYP154C1 and CYPs from antibiotic biosynthetic pathways. Protein Sci. Publ. Protein Soc. 2004, 13, 255–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Q.; Lamb, D.C.; Kelly, S.L.; Lei, L.; Guengerich, F.P. Cyclization of a cellular dipentaenone by Streptomyces coelicolor cytochrome P450 154A1 without oxidation/reduction. J. Am. Chem. Soc. 2010, 132, 15173–15175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Podust, L.M.; Kim, Y.; Arase, M.; Neely, B.A.; Beck, B.J.; Bach, H.; Sherman, D.H.; Lamb, D.C.; Kelly, S.L.; Waterman, M.R. The 1.92-A structure of Streptomyces coelicolor A3(2) CYP154C1. A new monooxygenase that functionalizes macrolide ring systems. J. Biol. Chem. 2003, 278, 12214–12221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, B.; Bellamine, A.; Lei, L.; Waterman, M.R. The role of Ile87 of CYP158A2 in oxidative coupling reaction. Arch Biochem. Biophys. 2012, 518, 127–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendes, M.V.; Anton, N.; Martin, J.F.; Aparicio, J.F. Characterization of the polyene macrolide P450 epoxidase from Streptomyces natalensis that converts de-epoxypimaricin into pimaricin. Biochem. J. 2005, 386, 57–62. [Google Scholar] [CrossRef] [Green Version]
- Xie, Z.; Niu, G.; Li, R.; Liu, G.; Tan, H. Identification and characterization of sanH and sanI involved in the hydroxylation of pyridyl residue during nikkomycin biosynthesis in Streptomyces ansochromogenes. Curr. Microbiol. 2007, 55, 537–542. [Google Scholar] [CrossRef]
- Chen, H.; Walsh, C.T. Coumarin formation in novobiocin biosynthesis: Beta-hydroxylation of the aminoacyl enzyme tyrosyl-S-NovH by a cytochrome P450 NovI. Chem. Biol. 2001, 8, 301–312. [Google Scholar] [CrossRef] [Green Version]
- Uhlmann, S.; Süssmuth, R.D.; Cryle, M.J. Cytochrome p450sky interacts directly with the nonribosomal peptide synthetase to generate three amino acid precursors in skyllamycin biosynthesis. ACS Chem. Biol. 2013, 8, 2586–2596. [Google Scholar] [CrossRef]
- Zhao, B.; Lei, L.; Vassylyev, D.G.; Lin, X.; Cane, D.E.; Kelly, S.L.; Yuan, H.; Lamb, D.C.; Waterman, M.R. Crystal structure of albaflavenone monooxygenase containing a moonlighting terpene synthase active site. J. Biol. Chem. 2009, 284, 36711–36719. [Google Scholar] [CrossRef] [Green Version]
- Takamatsu, S.; Lin, X.; Nara, A.; Komatsu, M.; Cane, D.E.; Ikeda, H. Characterization of a silent sesquiterpenoid biosynthetic pathway in Streptomyces avermitilis controlling epi-isozizaene albaflavenone biosynthesis and isolation of a new oxidized epi-isozizaene metabolite. Microb. Biotechnol. 2011, 4, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Moody, S.C.; Zhao, B.; Lei, L.; Nelson, D.R.; Mullins, J.G.; Waterman, M.R.; Kelly, S.L.; Lamb, D.C. Investigating conservation of the albaflavenone biosynthetic pathway and CYP170 bifunctionality in streptomycetes. FEBS J. 2012, 279, 1640–1649. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, H.; Omura, S. Avermectin Biosynthesis. Chem. Rev. 1997, 97, 2591–2610. [Google Scholar] [CrossRef] [PubMed]
- Lamb, D.C.; Ikeda, H.; Nelson, D.R.; Ishikawa, J.; Skaug, T.; Jackson, C.; Omura, S.; Waterman, M.R.; Kelly, S.L. Cytochrome p450 complement (CYPome) of the avermectin-producer Streptomyces avermitilis and comparison to that of Streptomyces coelicolor A3(2). Biochem. Biophys. Res. Commun. 2003, 307, 610–619. [Google Scholar] [CrossRef]
- Tetzlaff, C.N.; You, Z.; Cane, D.E.; Takamatsu, S.; Omura, S.; Ikeda, H. A gene cluster for biosynthesis of the sesquiterpenoid antibiotic pentalenolactone in Streptomyces avermitilis. Biochemistry 2006, 45, 6179–6186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Healy, F.G.; Krasnoff, S.B.; Wach, M.; Gibson, D.M.; Loria, R. Involvement of a cytochrome P450 monooxygenase in thaxtomin A biosynthesis by Streptomyces acidiscabies. J. Bacteriol. 2002, 184, 2019–2029. [Google Scholar] [CrossRef] [Green Version]
- Markowitz, V.M.; Chen, I.-M.A.; Palaniappan, K.; Chu, K.; Szeto, E.; Grechkin, Y.; Ratner, A.; Jacob, B.; Huang, J.; Williams, P. IMG: The integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 2011, 40, D115–D122. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef] [Green Version]
- Syed, K.; Mashele, S.S. Comparative analysis of P450 signature motifs EXXR and CXG in the large and diverse kingdom of fungi: Identification of evolutionarily conserved amino acid patterns characteristic of P450 family. PLoS ONE 2014, 9, e95616. [Google Scholar] [CrossRef] [Green Version]
- Syed, P.R.; Chen, W.; Nelson, D.R.; Kappo, A.P.; Yu, J.H.; Karpoormath, R.; Syed, K. Cytochrome P450 Monooxygenase CYP139 Family Involved in the Synthesis of Secondary Metabolites in 824 Mycobacterial Species. Int. J. Mol. Sci. 2019, 20, 2690. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Kuma, K.-I.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Boc, A.; Diallo, A.B.; Makarenkov, V. T-REX: A web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 2012, 40, W573–W579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howe, E.; Holton, K.; Nair, S.; Schlauch, D.; Sinha, R.; Quackenbush, J. Mev: Multiexperiment viewer. In Biomedical Informatics for Cancer Research; Springer: Berlin/Heidelberg, Germany, 2010; pp. 267–277. [Google Scholar]
- Blin, K.; Wolf, T.; Chevrette, M.G.; Lu, X.; Schwalen, C.J.; Kautsar, S.A.; Suarez Duran, H.G.; de Los Santos, E.L.C.; Kim, H.U.; Nave, M.; et al. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017, 45, W36–W41. [Google Scholar] [CrossRef] [PubMed]




| Species Name | P450s | No. F | No. SF | Species Name | P450s | No. F | No. SF |
|---|---|---|---|---|---|---|---|
| Streptomyces sp. Tu6071 | 22 | 13 | 20 | Streptomyces sp. CNT372 | 10 | 8 | 10 |
| Streptomyces purpureus KA281, ATCC 21405 | 22 | 17 | 20 | Streptomyces sp. CNS606 | 16 | 9 | 14 |
| Streptomyces sp. W007 | 28 | 12 | 24 | Streptomyces sp. 303MFCol5.2 | 23 | 14 | 22 |
| Streptomyces sp. TAA486-18 | 18 | 12 | 17 | Streptomyces acidiscabies 84-104 | 47 | 22 | 44 |
| Streptomyces lysosuperificus ATCC 31396 | 25 | 19 | 24 | Streptomyces roseosporus NRRL 11379 | 19 | 10 | 16 |
| Streptomyces sp. PVA 94-07 | 20 | 7 | 18 | Streptomyces sp. OspMP-M45 | 19 | 9 | 19 |
| Streptomyces sp. SPB78 | 20 | 12 | 20 | Streptomyces sp. AmelKG-A3 | 19 | 9 | 19 |
| Streptomyces canus 299MFChir4.1 | 28 | 17 | 27 | Streptomyces sp. S4 | 19 | 9 | 19 |
| Streptomyces sp. FxanaA7 | 30 | 15 | 29 | Streptomyces sp. SM8 | 18 | 8 | 16 |
| Streptomyces sulphureus DSM 40104 | 26 | 13 | 25 | Streptomyces sp. LaPpAH-199 | 26 | 11 | 21 |
| Streptomyces sp. MspMP-M5 | 44 | 20 | 41 | Streptomyces sp. 140Col2.1E | 22 | 9 | 17 |
| Streptomyces coelicoflavus ZG0656 | 17 | 12 | 16 | Streptomyces sp. DvalAA-21 | 24 | 10 | 22 |
| Streptomyces pristinaespiralis ATCC 25486 | 18 | 11 | 17 | Streptomyces sp. CNT371 | 17 | 13 | 17 |
| Streptomyces sp. LaPpAH-201 | 19 | 8 | 19 | Streptomyces somaliensis DSM 40738 | 10 | 8 | 9 |
| Streptomyces albulus CCRC 11814 | 64 | 26 | 50 | Streptomyces sp. 351MFTsu5.1 | 22 | 11 | 22 |
| Streptomyces viridochromogenes DSM 40736 | 24 | 15 | 24 | Streptomyces sp. DvalAA-83 | 24 | 10 | 22 |
| Streptomyces sp. LaPpAH-95 | 24 | 9 | 22 | Streptomyces sp. AmelKG-F2B | 24 | 17 | 23 |
| Streptomyces mirabilis YR139 | 42 | 26 | 41 | Streptomyces sp. CNT302 | 26 | 13 | 22 |
| Streptomyces sp. AA1529 | 26 | 15 | 24 | Streptomyces olindensis DAUFPE 5622 | 26 | 14 | 22 |
| Streptomyces atratus OK008 | 15 | 10 | 14 | Streptomyces sp. CNY243 | 17 | 14 | 16 |
| Streptomyces sp. PsTaAH-130 | 36 | 21 | 32 | Streptomyces sp. AA0539 | 19 | 10 | 19 |
| Streptomyces sp. CNT318 | 27 | 15 | 24 | Streptomyces atratus OK807 | 31 | 13 | 27 |
| Streptomyces sp. CNH099 | 16 | 12 | 16 | Streptomyces sp. CNS335 | 16 | 13 | 17 |
| Streptomyces sp. CNH287 | 16 | 12 | 16 | Streptomyces sp. FxanaC1 | 27 | 15 | 24 |
| Streptomyces sp. MnatMP-M77 | 32 | 14 | 27 | Streptomyces sp. WMMB 322 | 19 | 11 | 17 |
| Streptomyces zinciresistens K42 | 19 | 11 | 18 | Streptomyces sp. TOR3209 | 20 | 13 | 19 |
| Streptomyces sp. So1WspMP-so12th | 22 | 11 | 19 | Streptomyces sp. AmelKG-E11A | 24 | 15 | 22 |
| Streptomyces sp. GXT6 | 13 | 8 | 11 | Streptomyces sp. PP-C42 | 16 | 6 | 14 |
| Streptomyces roseosporus NRRL 15998 | 19 | 10 | 16 | Streptomyces sp. DpondAA-E10 | 25 | 10 | 22 |
| Streptomyces sp. LaPpAH-108 | 24 | 12 | 23 | Streptomyces sp. HPH0547 | 32 | 18 | 32 |
| Streptomyces aurantiacus JA 4570 | 30 | 20 | 30 | Streptomyces sp. DpondAA-A50 | 25 | 10 | 22 |
| Streptomyces hygroscopicus ATCC 53653 | 57 | 21 | 49 | Streptomyces sp. TAA040 | 15 | 10 | 15 |
| Streptomyces sp. Tu 6176 | 30 | 15 | 26 | Streptomyces sp. PgraA7 | 23 | 10 | 20 |
| Streptomyces ghanaensis ATCC 14672 | 35 | 20 | 34 | Streptomyces sp. FxanaD5 | 15 | 11 | 15 |
| Streptomyces sp. KhCrAH-337 | 26 | 12 | 22 | Streptomyces sp. LamerLS-316 | 25 | 11 | 22 |
| Streptomyces sp. LaPpAH-202 | 19 | 8 | 19 | Streptomyces viridochromogenes Tue57 | 31 | 17 | 29 |
| Streptomyces sp. UNC401CLCol | 15 | 11 | 15 | Streptomyces sp. GBA 94-10 | 20 | 7 | 18 |
| Streptomyces sp. SirexAA-H | 21 | 12 | 20 | Streptomyces sp. CNQ-525 | 18 | 14 | 18 |
| Streptomyces turgidiscabies Car8 | 28 | 20 | 27 | Streptomyces sp. SceaMP-e96 | 41 | 18 | 36 |
| Streptomyces sp. KhCrAH-40 | 26 | 12 | 22 | Streptomyces mirabilis OK461 | 37 | 16 | 31 |
| Streptomyces rimosus rimosus ATCC 10970 | 54 | 30 | 52 | Streptomyces sp. LaPpAH-185 | 44 | 27 | 40 |
| Streptomyces gancidicus BKS 13-15 | 18 | 11 | 17 | Streptomyces exfoliatus DSMZ 41693 | 26 | 16 | 24 |
| Streptomyces auratus AGR0001 | 35 | 14 | 33 | Streptomyces sp. PsTaAH-137 | 29 | 16 | 28 |
| Kitasatospora sp. SolWspMP-SS2h | 25 | 15 | 24 | Streptomyces sp. Amel2xE9 | 27 | 15 | 26 |
| Streptomyces sp. NTK 937 | 17 | 8 | 17 | Streptomyces sp. AmelKG-D3 | 22 | 11 | 19 |
| Streptomyces sp. ScaeMP-e48 | 19 | 10 | 17 | Streptomyces prunicolor NBRC 13075 | 44 | 18 | 39 |
| Streptomyces sp. HmicA12 | 25 | 14 | 24 | Streptomyces sp. e14 | 28 | 13 | 25 |
| Streptomyces griseoaurantiacus M045 | 16 | 11 | 16 | Streptomyces sp. CNX435 | 12 | 9 | 12 |
| Streptomyces afghaniensis 772 | 28 | 17 | 29 | Streptomyces sp. HCCB10043 | 17 | 10 | 14 |
| Streptomyces sulphureus L180 | 19 | 11 | 19 | Streptomyces sp. JS01 | 24 | 11 | 19 |
| Streptomyces sp. KhCrAH-340 | 26 | 12 | 22 | Streptomyces chartreusis NRRL 3882 | 29 | 19 | 26 |
| Streptomyces sp. C | 30 | 17 | 27 | Streptomyces sp. CNY228 | 19 | 9 | 19 |
| Streptomyces violaceusniger SPC6 | 13 | 8 | 12 | Streptomyces sp. Amel2xB2 | 27 | 13 | 25 |
| Streptomyces sp. HGB0020 | 23 | 13 | 22 | Streptomyces sp. LaPpAH-165 | 24 | 9 | 22 |
| Streptomyces sp. CNS615 | 27 | 15 | 24 | Streptomyces albulus ZPM | 68 | 27 | 51 |
| Streptomyces tsukubaensis NRRL 18488 | 30 | 18 | 30 | Streptomyces albulus NK660 | 64 | 27 | 50 |
| Streptomyces vitaminophilus DSM 41686 | 18 | 10 | 15 | Streptomyces noursei | 64 | 26 | 52 |
| Streptomyces sp. SA3_actG | 21 | 12 | 20 | Streptomyces violaceusniger Tu4113 | 50 | 16 | 42 |
| Streptomyces bottropensis ATCC 25435 (2517572239) | 31 | 19 | 30 | Streptomyces bingchenggensis | 49 | 26 | 44 |
| Streptomyces sp. CNQ865 | 16 | 13 | 16 | Streptomyces rapamycinicus | 63 | 23 | 56 |
| Streptomyces sp. CNT360 | 19 | 13 | 18 | Streptomyces sp. 769 | 59 | 24 | 49 |
| Streptomyces sp. 142MFCol3.1 | 27 | 14 | 24 | Streptomyces hygroscopicus subsp. jinggangensis 5008 | 38 | 18 | 33 |
| Streptomyces sp. ScaeMP-e122 | 25 | 11 | 23 | Streptomyces cattleya NRRL 8058 = DSM 46488 | 41 | 21 | 38 |
| Streptomyces griseoflavus Tu4000 | 20 | 15 | 19 | Streptomyces cattleya NRRL 8057 | 40 | 20 | 37 |
| Streptomyces sp. ACT-1 | 30 | 13 | 26 | Streptomyces hygroscopicus subsp. jinggangensis TL01 | 37 | 18 | 33 |
| Streptomyces sp. TAA204 | 18 | 10 | 16 | Streptomyces avermitilis MA-4680 | 52 | 23 | 42 |
| Streptomyces sp. SPB74 | 18 | 10 | 18 | Streptomyces collinus | 34 | 16 | 27 |
| Streptomyces sp. CNQ329 | 13 | 10 | 13 | Streptomyces lydicus A02 | 38 | 19 | 35 |
| Streptomyces sp. 4F | 16 | 11 | 15 | Streptomyces lydicus 103 | 32 | 13 | 29 |
| Streptomyces sp. KhCrAH-244 | 26 | 12 | 22 | Streptomyces sp. Mg1 | 37 | 21 | 36 |
| Streptomyces chartreusis NRRL 12338 | 23 | 15 | 23 | Streptomyces leeuwenhoekii C34(2013) | 36 | 17 | 34 |
| Streptomyces sviceus ATCC 29083 | 19 | 12 | 19 | Streptomyces pratensis/flavogriseus IAF 45 | 29 | 16 | 26 |
| Streptomyces sp. CcalMP-8W | 23 | 12 | 20 | Streptomyces reticuli | 47 | 26 | 43 |
| Streptomyces sp. SS | 15 | 11 | 15 | Streptomyces griseus | 28 | 13 | 24 |
| Streptomyces sp. CNQ766 | 16 | 13 | 16 | Streptomyces sp. PAMC 26508 | 29 | 16 | 26 |
| Streptomyces sp. URHA0041 | 16 | 9 | 15 | Streptomyces sp. SirexAA-E | 24 | 10 | 22 |
| Streptomyces sp. CNB091 | 27 | 14 | 24 | Streptomyces davawensis | 32 | 19 | 30 |
| Streptomyces flavidovirens DSM 40150 | 24 | 15 | 23 | Streptomyces cyaneogriseus | 30 | 14 | 28 |
| Streptomyces yeochonensis CN732 | 18 | 11 | 18 | Streptomyces lincolnensis | 24 | 15 | 23 |
| Streptomyces viridosporus T7A, ATCC 39115 | 32 | 19 | 31 | Streptomyces pristinaespiralis HCCB 10218 | 23 | 12 | 18 |
| Streptomyces sp. FXJ7.023 | 27 | 12 | 23 | Streptomyces venezuelae | 23 | 16 | 21 |
| Streptomyces mirabilis OV308 | 28 | 14 | 27 | Streptomyces sp. CFMR 7 | 24 | 13 | 20 |
| Streptomyces sp. AW19M42 | 27 | 12 | 24 | Streptomyces vietnamensis | 30 | 20 | 29 |
| Streptomyces sp. ATexAB-D23 | 28 | 11 | 26 | Streptomyces xiamenensis 318 | 19 | 12 | 19 |
| Streptomyces sp. BoleA5 | 17 | 8 | 15 | Streptomyces coelicolor | 18 | 10 | 17 |
| Streptomyces sp. AA4 | 35 | 17 | 29 | Streptomyces albus J1074 | 18 | 9 | 18 |
| Streptomyces sp. CNS654 | 27 | 10 | 22 | Streptomyces ambofaciens | 19 | 10 | 18 |
| Streptomyces ipomoeae 91-03 | 44 | 26 | 43 | Streptomyces lividans | 20 | 10 | 18 |
| Streptomyces sp. DpondAA-B6 | 19 | 9 | 19 | Streptomyces scabiei 87.22 | 30 | 16 | 30 |
| Streptomyces sp. PCS3-D2 | 25 | 18 | 24 | Streptomyces glaucescens | 18 | 11 | 17 |
| Streptomyces sp. PRh5 | 57 | 20 | 51 | Streptomyces albus DSM 41398 | 25 | 13 | 24 |
| Streptomyces sp. CNR698 | 29 | 17 | 26 | Streptomyces fulvissimus | 19 | 10 | 16 |
| Amycolatopsis sp. 75iv2, ATCC 39116 | 28 | 18 | 27 | Streptomyces sp. CNQ-509 | 16 | 11 | 16 |
| Streptomyces cattleya ATCC 35852 | 41 | 21 | 38 | Streptomyces rubrolavendulae | 20 | 12 | 19 |
| Streptomyces sp. WMMB 714 | 21 | 10 | 18 | Streptomyces clavuligerus | 64 | 30 | 58 |
| Streptomyces scabrisporus DSM 41855 | 37 | 27 | 36 | Streptomyces griseochromogenes | 46 | 24 | 40 |
| Streptomyces sp. Ncost-T6T-1 | 25 | 14 | 22 | Streptomyces sp. S10(2016) | 20 | 15 | 20 |
| Streptomyces sp. CNB632 | 16 | 12 | 16 | Streptomyces globisporus | 23 | 13 | 19 |
| Streptomyces mobaraensis NBRC 13819 | 22 | 13 | 21 | Streptomyces sp. CdTB01 | 26 | 17 | 25 |
| Streptomyces sp. KhCrAH-43 | 26 | 12 | 22 | Streptomyces parvulus | 25 | 15 | 25 |
| Streptomyces sp. PsTaAH-124 | 32 | 16 | 27 | Streptomyces sp. SAT1 | 25 | 15 | 22 |
| Streptomyces sp. Amel2xC10 | 15 | 10 | 15 |
| Streptomyces Species | Mycobacterial Species | Bacillus Species | Cyanobacterial Species | |
|---|---|---|---|---|
| Total No. of species analyzed | 203 | 60 | 128 | 114 |
| No. of P450s | 5460 | 1784 | 507 | 341 |
| No. of families | 253 | 77 | 13 | 36 |
| No. of subfamilies | 698 | 132 | 28 | 79 |
| Dominant P450 family | CYP107 | CYP125 | CYP107 | CYP110 |
| Average no. of P450s | 27 | 30 | 4 | 3 |
| No. of BGCs * | 4457 | 898 | 1098 | 770 |
| Average no. of BGCs | 31 | 15 | 9 | 7 |
| No. of P450s part of BGCs | 1231 | 204 | 112 | 27 |
| Percentage of P450s part of BGCs | 22 | 11 | 22 | 8 |
| Reference | This work | [20,21] | [22] | [23] |
| P450 Family | Dominant Subfamilies | ||||||
|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | |
| CYP157 | 174 | 177 | |||||
| CYP154 | 127 | 164 | 76 | ||||
| CYP156 | 120 | ||||||
| CYP102 | 78 | 48 | |||||
| CYP159 | 125 | ||||||
| CYP125 | 104 | ||||||
| CYP147 | 73 | ||||||
| CYP158 | 91 | ||||||
| CYP1035 | 79 | ||||||
| CYP163 | 50 | ||||||
| CYP180 | 54 | ||||||
| CYP170 | 57 | ||||||
| CYP124 | 50 | ||||||
| CYP1047 | 43 | ||||||
| CYP152 | 42 | ||||||
| CYP251 | 23 | ||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mnguni, F.C.; Padayachee, T.; Chen, W.; Gront, D.; Yu, J.-H.; Nelson, D.R.; Syed, K. More P450s Are Involved in Secondary Metabolite Biosynthesis in Streptomyces Compared to Bacillus, Cyanobacteria, and Mycobacterium. Int. J. Mol. Sci. 2020, 21, 4814. https://doi.org/10.3390/ijms21134814
Mnguni FC, Padayachee T, Chen W, Gront D, Yu J-H, Nelson DR, Syed K. More P450s Are Involved in Secondary Metabolite Biosynthesis in Streptomyces Compared to Bacillus, Cyanobacteria, and Mycobacterium. International Journal of Molecular Sciences. 2020; 21(13):4814. https://doi.org/10.3390/ijms21134814
Chicago/Turabian StyleMnguni, Fanele Cabangile, Tiara Padayachee, Wanping Chen, Dominik Gront, Jae-Hyuk Yu, David R. Nelson, and Khajamohiddin Syed. 2020. "More P450s Are Involved in Secondary Metabolite Biosynthesis in Streptomyces Compared to Bacillus, Cyanobacteria, and Mycobacterium" International Journal of Molecular Sciences 21, no. 13: 4814. https://doi.org/10.3390/ijms21134814
APA StyleMnguni, F. C., Padayachee, T., Chen, W., Gront, D., Yu, J. -H., Nelson, D. R., & Syed, K. (2020). More P450s Are Involved in Secondary Metabolite Biosynthesis in Streptomyces Compared to Bacillus, Cyanobacteria, and Mycobacterium. International Journal of Molecular Sciences, 21(13), 4814. https://doi.org/10.3390/ijms21134814