1. Introduction
Wheat (
Triticum aestivum L.) is one of the most important food crops worldwide, as it provides humans most of their daily calories and proteins. With the growing of the human population, wheat demand is expected to increase by 66% in 2040 [
1]. However, wheat production has been continually threatened by abiotic and biotic stresses. Stripe rust (also known as yellow rust), caused by fungal pathogen
Puccinia striiformis Westend. f. sp.
tritici Erikss. (
Pst), is especially a devastating disease in the world [
2,
3], and the fungus is ranked among the global burden pathogens and pests of major crops [
4]. The disease can cause more than 90% yield losses in fields grown with susceptible cultivars if the weather conditions are extremely favorable to stripe rust development [
5].
Growing resistant cultivars and application of fungicides are two major approaches for control of stripe rust. Use of fungicides adds significant cost to production; has potential to select fungicide-tolerant populations of the pathogen; is potentially harmful to humans, animals, and the environment; and therefore, is limited in organic farming and areas under protection for wildlife habitats. In contrast, growing resistant cultivars does not have these problems, but is easy to use, efficient, and more effective for control of stripe rust [
6,
7]. On the other hand, resistance may have some other problems. For example, resistant cultivars may become susceptible to new virulent races of the pathogen or the resistance level may not be adequate when the disease is severe. However, these problems can be solved by developing cultivars with durable, high-level resistance [
5].
Resistance to stripe rust is generally described in two types: all-stage resistance and high-temperature adult-plant (HTAP) resistance [
2,
7]. All-stage resistance (ASR), also called seedling resistance, is normally controlled by a single gene that provides high-level resistance throughout the growth stages. However, ASR is usually race-specific, and cultivars with this type of resistance can be easily overcome by new virulent races of the pathogen. More than 300
Pst races have been identified from wheat stripe rust samples collected from the US and several countries, and the recent predominant races in the US are virulent on 16 of the 18
Yr single-gene lines in the Avocet background that are used to differentiate
Pst races [
8,
9,
10]. In contrast, HTAP resistance, which is expressed when the plant grows old and weather becomes warm, is nonrace-specific and durable [
7,
11]. The quantitative nature makes HTAP resistance breeding relatively difficult compared to ASR, and also often the partial resistance may not provide adequate control if the disease starts in the early growth stage and the weather conditions favor the disease but do not favor expression of HTAP resistance. Therefore, it is better to combine both types of resistance by pyramiding genes for effective ASR and HTAP resistance to utilize their advantages and overcome the disadvantages. This approach requires a large number of genes with user-friendly markers for marker-assisted selection (MAS). Thus, mapping new resistance genes is a continual task for developing wheat cultivars with durable, high-level resistance to stripe rust to achieve sustainable and more effective control.
Wheat landraces are valuable sources of resistance to stripe rust. PI 181410, a spring wheat landrace originally from Afghanistan (
https://npgsweb.ars-grin.gov/gringlobal/accessiondetail.aspx?id=1157002), has shown excellent resistance to stripe rust [
12]. However, the genes for resistance to stripe rust in PI 181410 were not known. Therefore, the objectives of the present study were to (1) characterize stripe rust resistance in PI 181410, (2) identify and map genes or quantitative trait loci (QTL) conferring resistance, and (3) develop molecular markers for marker-assisted selection.
3. Discussion
Wheat landrace PI 181410 has been consistently showing high-level resistance to stripe rust in the US Pacific Northwest region since 2006. The four-way tests conducted with predominant and most virulent races of Pst in both seedling and adult-plant stages at both LT and HT profiles showed that PI 181410 has both race-specific ASR and high-level HTAP resistance. Using the high-density genetic maps and multiple environmental tests for QTL mapping, we identified three QTL in PI 181410 for ASR, QYrPI181410.wgp-4AS, QYrPI181410.wgp-4BL, and QYrPI181410.wgp-5BL.1. In addition to the QTL on 4AS, we identified three more QTL, designated as QYrPI181410.wgp-1BL, QYrPI181410.wgp-5AS, and QYrPI181410.wgp-5BL.2 in PI 181410 conferring resistance observed at the adult-plant stage in the fields. Interestingly, we also detected two QTL, designated as QYrAvS.wgp-2BS and QYrAvS.wgp-2BL, from AvS with minor effects only on IT at Mount Vernon and Pullman, respectively. Each of the QTL were located within a relatively small genomic region.
QYrPI181410.wgp-1BL associated with minor HTAP resistance was detected at the distal end of the long arm of 1B.
Yr29 is also located at the distal end of 1BL [
13]. At the
Yr29 location, eleven QTL were identified with different phenotypic effects [
14]. Six of the eleven QTL have been reported to provide significant effects on stripe rust resistance in multiple environmental tests, such as
QYr.sun-1B,
QYr.jic-1B,
QYr.tam-1B,
QYr.cim-1BL,
QYr-1B,
QYr.cim-1BL, and
QYr.1BL [
15,
16,
17,
18,
19,
20]. Other QTL including
QYrPst.jic-1B were reported to have minor effects on stripe rust resistance [
14,
21]. Based on the chromosome regions,
QYrPI181410.wgp-1BL is likely
Yr29. The present study and previous studies suggests that the long arm of chromosome 1B carries multiple genes for resistance to stripe rust.
Two minor-effect QTL,
QYrAvS.wgp-2BS and
QYrAvS.wgp-2BL, were identified from susceptible parent AvS.
QYrPI181410.wgp-2BS was mapped close to the distal end of the short arm. According to previous studies, two QTL including
QYr.inra-2BS [
22] and
QYr.wgp-2B.1 [
23] were mapped at the same region as
QYrAvS.wgp-2BS.
QYr.inra-2BS contributed moderate resistance to stripe rust at both seedling and adult-plant stages [
22]. Identified in several US Pacific Northwest wheat cultivars by genome-wide association mapping,
QYr.wgp-2B.1 contributed adult-plant resistance to stripe rust and was significantly associated with both IT and SE in several environments including Pullman and Mount Vernon, Washington [
23]. However,
QYrPI181410.wgp-2BS was only associated with IT in Mount Vernon. Thus,
QYrAvS.wgp-2BS is likely a locus different from
QYr.inra-2BS and
QYr.wgp-2B.1.
QYrAvS.wgp-2BL was only detected using one set of IT data in a single field condition in Pullman. It was located at the distal end of 2BL. Rosewarne et al. [
17] reported a minor QTL affecting adult-plant resistance in AvS flanked by simple sequence repeat (SSR) markers
Xgwm1027 and
Xgwm619 at the same location on chromosome 2BL as
QYrAvS.wgp-2BL. Since their QTL and
QYrAvS.wgp-2BL were detected in the same wheat genotype in the same chromosomal location, they should be the same gene.
QYrPI181410.wgp-5AS had a minor effect on reducing disease severity in PI 181410 and was located at the telomere of chromosome 5AS. As no stripe rust resistance QTL have been reported at this location, the 5AS QTL is a new QTL for resistance to stripe rust.
Two QTL,
QYrPI181410.wgp-5BL.1 and
QYrPI181410.wgp-5BL.2, from PI 181410 were mapped to different locations on chromosome 5BL. These QTL conferred resistance to stripe rust at different plant growth stages.
QYrPI181410.wgp-5BL.1, associated with ASR, was located at the middle region of 5BL, while
QYrPI181410.wgp-5BL.2, associated with HTAP resistance, was mapped to the distal end of 5BL. Five previously reported QTL, including
QYr.caas-5BL.1 from Libellula [
24],
QYrdr.wgp-5BL.2 [
25],
QYrco.wpg-5B [
26],
QYr.jirc-5BL [
27], and
QYr.inra-5BL.1, were at the chromosomal regions similar to
QYrPI181410.wgp-5BL.1 based on their flanking markers. All these QTL exhibit adult-plant resistance to multiple races, however,
QYrPI181410.wgp-5BL.1 conferred ASR to races PSTv-4 and PSTv-14. Based on the different types of resistance,
QYrPI181410.wgp-5BL.1 may be different from those known QTL, but further work is needed to determine their genetic relationships. Two QTL,
QYr.caas-5BL.2 flanked by
Xbarc142 and
Xgwm604 [
24] and
QYr.caas- 5BL.3 flanked by SSR markers
Xwmc75 and
Xbarc275 [
28], were mapped to the distal end of 5BL [
14]. Compared with the consensus map [
29],
QYrPI181410.wgp-5BL.2 should be close to the far distal end of the 5BL and not in the same region as the above two QTL. Thus,
QYrPI181410.wgp-5BL.2 is likely a new QTL for stripe rust resistance.
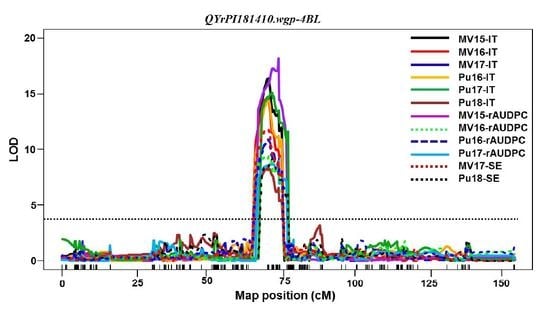
Among the QTL identified in PI 181410,
QYrPI181410.wgp-4BL was the most effective. It was consistently detected using all 16 sets of phenotypic data collected from all field tests and both seedling and adult-plant tests in the greenhouse. Its effect was much stronger in the adult-plant stage in the fields and at HT in the greenhouse test than the seedling stage at LT, and therefore, it should be considered as a HTAP resistance gene.
QYrPI181410.wgp-4BL was mapped to an interval of 9.0 cM (67.0~76.0 cM) close to the centromeric region of 4BL according to the consensus map [
29,
30]. The overlap of the genomic regions detected by all 16 data sets narrowed the QTL to 4.0 cM between 68.0 and 72.0 cM positions (
Table 5,
Figure 4). In previous studies,
Yr50 and
Yr62 were also mapped to the same location on 4BL as
QYrPI181410.wgp-4BL. The dominant resistance gene,
Yr50, derived from
Thinopyrum intermedium, was mapped on 4BL and flanked by SSR markers
Xbarc1096 and
Xwmc47, and
Yr50 exhibited ASR to all tested Chinese
Pst races and eight US predominant
Pst races including PSTv-40 [
31,
32]. In contrast,
QYrPI181410.wgp-4BL is from common wheat landrace PI 181410 and confers strong HTAP resistance with only a minor effect at seedling stage. The differences clearly indicate that
QYrPI181410.wgp-4BL is different from
Yr50.
Yr62, from Portuguese spring wheat variety PI 192252, was flanked by SSR markers
Xgwm192 and
Xgwm251 on 4BL [
32]. This gene provides a higher level HTAP resistance (40%–60% of phenotypic variation) than
QYrPI181410.wgp-4BL (29.2%–46.9%).
Yr62 is not effective at the seedling stage. Thus,
QYrPI181410.wgp-4BL is likely different from
Yr62. Further studies are needed to determine their relationship using the allelism test approach.
Two major QTL,
QPst.jic-4B and
QYrva.vt-4BL, were also mapped around the centromeric region on 4BL [
21,
23].
QPst.jic-4B, identified from a German winter wheat cultivar Alcedo, was flanked by amplified fragment length polymorphism (AFLP) marker
S24M37_130 and SSR marker
Xcfd39 and peaked at
Xwmc692 on 4BL [
21].
QYrva.vt-4BL, identified from a soft red winter wheat VA00W-38, was flanked by SSR markers
Xwmc692 and
Xgwm149 on 4BL [
33]. Both
QPst.jic-4B and
QYrva.vt-4BL explained less effect (17.9%–28.9% and 19.3%, respectively) of the phenotypic variation. Thus,
QYrPI181410.wgp-4BL is unlikely the same as any of these two QTL in winter wheat cultivars. As
QYrPI181410.wgp-4BL is the most useful among the QTL detected in the present study, we developed three KASP markers for this QTL. The KASP markers performed in the same way as the original SNP markers between the parents and among the RILs, and they were highly polymorphic when tested with 155 wheat cultivars and breeding lines from the US Pacific Northwest. None of the wheat materials have the
KIWB80 allele or the PI 181410 haplotype of the three markers. Therefore, the KASP marker
KIWB80 or the combination of the three markers can be used in MAS for incorporating the effective QTL into various wheat backgrounds for developing new cultivars with durable, high-level resistance to stripe rust.