Self-Targeting of Carbon Dots into the Cell Nucleus: Diverse Mechanisms of Toxicity in NIH/3T3 and L929 Cells
Abstract
:1. Introduction
2. Results
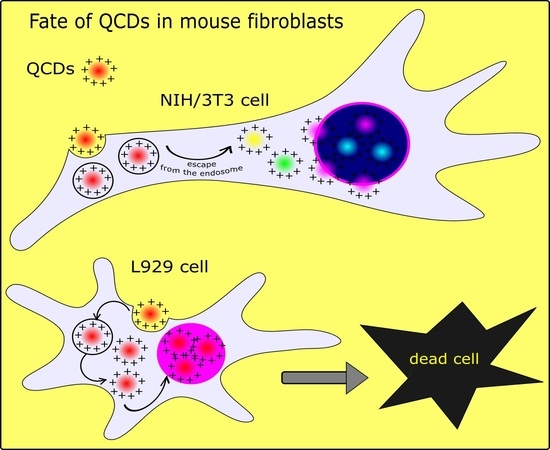
2.1. Detection of QCDs Inside Nuclei, Viability and DNA Damage
2.1.1. Results of QCD-Labeled NIH/3T3 Cells
2.1.2. Results of QCD-Labeled L929 Cells
2.2. Concentration Dependent Uptake and Endocytosis of Both Cell Lines
2.3. Cell Cycle Analysis of Both Lines
3. Materials and Methods
3.1. Carbon Dots
3.2. Cell Cultivation
3.3. Fluorescence Microspectroscopy
3.4. Cell Cycle, Concentration Dependent Uptake, Endocytosis
3.5. Viability
3.6. Comet Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, H.; Bai, Y.; Zhou, Y.; Feng, C.; Liu, L.; Fang, L.; Liang, J.; Xiao, S. Blue and cyan fluorescent carbon dots: One-pot synthesis, selective cell imaging and their antiviral activity. RSC Adv. 2017, 7, 28016–28023. [Google Scholar] [CrossRef] [Green Version]
- Georgakilas, V.; Perman, J.A.; Tucek, J.; Zboril, R. Broad family of carbon nanoallotropes: Classification, chemistry, and applications of fullerenes, carbon dots, nanotubes, graphene, nanodiamonds, and combined superstructures. Chem. Rev. 2015, 115, 4744–4822. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.Y.; Shen, W.; Gao, Z.Q. Carbon quantum dots and their applications. Chem. Soc. Rev. 2015, 44, 362–381. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hu, A. Carbon quantum dots: Synthesis, properties and applications. J. Mater. Chem. C 2014, 2, 6921–6939. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Yang, J.; Jia, L.; Yu, J.S. Ethanol in aqueous hydrogen peroxide solution: Hydrothermal synthesis of highly photoluminescent carbon dots as multifunctional nanosensors. Carbon 2015, 93, 999–1007. [Google Scholar] [CrossRef]
- Zhai, Y.L.; Zhu, Z.J.; Zhu, C.Z.; Ren, J.T.; Wang, E.K.; Dong, S.J. Multifunctional water-soluble luminescent carbon dots for imaging and Hg2+ sensing. J. Mater. Chem. B 2014, 2, 6995–6999. [Google Scholar] [CrossRef]
- Shi, W.; Li, X.H.; Ma, H.M. A tunable ratiometric pH sensor based on carbon nanodots for the quantitative measurement of the intracellular pH of whole cells. Angew. Chem. Int. Ed. 2012, 51, 6432–6435. [Google Scholar] [CrossRef]
- Feng, J.; Wang, W.J.; Hai, X.; Yu, Y.L.; Wang, J.H. Green preparation of nitrogen-doped carbon dots derived from silkworm chrysalis for cell imaging. J. Mater. Chem. B 2016, 4, 387–393. [Google Scholar] [CrossRef]
- Yu, H.; Zhao, Y.; Zhou, C.; Shang, L.; Peng, Y.; Cao, Y.; Wu, L.Z.; Tung, C.H.; Zhang, T.R. Carbon quantum dots/TiO2 composites for efficient photocatalytic hydrogen evolution. J. Mater. Chem. A 2014, 2, 3344–3351. [Google Scholar] [CrossRef]
- Fernando, K.A.S.; Sahu, S.; Liu, Y.M.; Lewis, W.K.; Guliants, E.A.; Jafariyan, A.; Wang, P.; Bunker, C.E.; Sun, Y.P. Carbon quantum dots and applications in photocatalytic energy conversion. ACS Appl. Mater. Interfaces 2015, 7, 8363–8376. [Google Scholar] [CrossRef]
- Pan, J.; Sheng, Y.; Zhang, J.; Wei, J.; Huang, P.; Zhang, X.; Feng, B. Preparation of carbon quantum dots/TiO2 nanotubes composites and their visible light catalytic applications. J. Mater. Chem. A 2014, 2, 18082–18086. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Y.; Wang, Y.; Kalytchuk, S.; Kershaw, S.; Wang, Y.; Wang, P.; Zhang, T.; Zhao, Y.; Zhang, H.; et al. Color-switchable electroluminescence of carbon dot light-emitting diodes. ACS Nano 2013, 7, 11234–11241. [Google Scholar] [CrossRef]
- Batmunkh, M.; Shearer, C.J.; Biggs, M.J.; Shapter, J.G. Nanocarbons for mesoscopic perovskite solar cells. J. Mater. Chem. A 2015, 3, 9020–9031. [Google Scholar] [CrossRef] [Green Version]
- Wei, Z.; Yan, K.; Chen, H.; Yi, Y.; Zhang, T.; Long, X.; Li, J.; Zhang, L.; Wang, J.; Yang, S. Cost-efficient clamping solar cells using candle soot for hole extraction from ambipolar perovskites. Energy Environ. Sci. 2014, 7, 3326–3333. [Google Scholar] [CrossRef]
- Cai, Q.Y.; Li, J.; Ge, J.; Zhang, L.; Hu, Y.L.; Li, Z.H.; Qu, L.B. A rapid fluorescence “switch-on” assay for glutathione detection by using carbon dots-MnO2 nanocomposites. Biosens. Bioelectron. 2015, 72, 31–36. [Google Scholar] [CrossRef]
- Pandey, S.; Thakur, M.; Mewada, A.; Anjarlekar, D.; Mishra, N.; Sharon, M. Carbon dots functionalized gold nanorod mediated delivery of doxorubicin: Tri-functional nano-worms for drug delivery, photothermal therapy and bioimaging. J. Mater. Chem. B 2013, 1, 4972–4982. [Google Scholar] [CrossRef]
- Huang, P.; Lin, J.; Wang, X.; Wang, Z.; Zhang, C.; He, M.; Wang, K.; Chen, F.; Li, Z.; Shen, G.; et al. Light-triggered theranostics based on photosensitizer-conjugated carbon dots for simultaneous enhanced-fluorescence imaging and photodynamic therapy. Adv. Mater. 2012, 24, 5104–5110. [Google Scholar] [CrossRef] [Green Version]
- Mewada, A.; Pandey, S.; Thakur, M.; Jadhav, D.; Sharon, M. Swarming carbon dots for folic acid mediated delivery of doxorubicin and biological imaging. J. Mater. Chem. B 2014, 2, 698–705. [Google Scholar] [CrossRef]
- Kim, J.; Park, J.; Kim, H.; Singha, K.; Kim, W.J. Transfection and intracellular properties of carbon dot-gold nanoparticle molecular assembly conjugated with PEI-pDNA. Biomaterials 2013, 34, 7168–7180. [Google Scholar] [CrossRef]
- Liu, C.J.; Zhang, P.; Zhai, X.Y.; Tian, F.; Li, W.C.; Yang, J.H.; Liu, Y.; Wang, H.B.; Wang, W.; Liu, W.G. Nano-carrier for gene delivery and bioimaging based on carbon dots with PEI-passivation enhanced fluorescence. Biomaterials 2012, 33, 3604–3613. [Google Scholar] [CrossRef]
- Shao, D.; Lu, M.; Xu, D.; Zheng, X.; Pan, Y.; Song, Y.; Xu, J.; Li, M.; Zhang, M.; Li, J.; et al. Carbon dots for tracking and promoting the osteogenic differentiation of mesenchymal stem cells. Biomater. Sci. 2017, 5, 1820. [Google Scholar] [CrossRef] [PubMed]
- Pasinszki, T.; Krebsz, M.; Tung, T.T.; Losic, D. Carbon nanomaterial based biosensors for non-invasive detection of cancer and disease biomarkers for clinical diagnosis. Sensors 2017, 17, 1919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerrero, E.D.; Lopez-Velazquez, A.M.; Ahlawat, J.; Narayan, M. Carbon Quantum Dots for Treatment of Amyloid Disorders. ACS Appl. Nano Mater. 2021, 4, 2423–2433. [Google Scholar] [CrossRef] [PubMed]
- Pirsaheba, M.; Mohammadi, S.; Salimi, A. Current advances of carbon dots based biosensors for tumor marker detection, cancer cells analysis and bioimaging. TrAC Trends Anal. Chem. 2019, 115, 83–99. [Google Scholar] [CrossRef]
- Ghosh, S.; Ghosal, K.; Mohammad, S.A.; Sarkar, K. Dendrimer functionalized carbon quantum dot for selective detection of breast cancer and gene therapy. Chem. Eng. J. 2019, 373, 468–484. [Google Scholar] [CrossRef]
- Mohammadi, S.; Salimi, A.; Hamd-Ghadareh, S.; Fathi, F.; Soleimani, F. A FRET immunosensor for sensitive detection of CA 15-3 tumor marker in human serum sample and breast cancer cells using antibody functionalized luminescent carbon-dots and AuNPs-dendrimer aptamer as donor-acceptor pair. Anal. Biochem. 2018, 557, 18–26. [Google Scholar] [CrossRef]
- Ehtesabi, H.; Amirfazli, M.; Massah, F.; Bagheri, Z. Application of functionalized carbon dots in detection, diagnostic, disease treatment, and desalination: A review. Adv. Nat. Sci. Nanosci. Nanotechnol. 2020, 11, 025017. [Google Scholar] [CrossRef]
- Sun, Y.P.; Zhou, B.; Lin, Y.; Wang, W.; Fernando, K.A.S.; Pathak, P.; Meziani, M.J.; Harruff, B.A.; Wang, X.; Wang, H.F.; et al. Quantum-sized carbon dot for bright and colorful photoluminescence. J. Am. Chem. Soc. 2006, 128, 7756–7757. [Google Scholar] [CrossRef]
- Hu, Y.P.; Yang, J.; Tian, J.W.; Jia, L.; Yu, J.S. Waste frying oil as a precursor for one-step synthesis of sulfur-doped carbon dots with pH-sensitive photoluminescence. Carbon 2014, 77, 775–782. [Google Scholar] [CrossRef]
- Bao, L.; Zhang, Z.L.; Tian, Z.Q.; Zhang, L.; Liu, C.; Lin, Y.; Qi, B.P.; Pang, D.W. Electrochemical tuning of luminescent carbon nanodots: From preparation to luminescence mechanism. Adv. Mater. 2011, 23, 5801–5806. [Google Scholar] [CrossRef]
- Algarra, M.; Perez-Martin, M.; Cifuentes-Rueda, M.; Jimenez-Jimenez, J.; Esteves da Silva, J.C.G.; Bandosz, T.J.; Rodriguez-Castellon, E.; Lopez Navarrete, J.T.; Casado, J. Carbon dots obtained using hydrothermal treatment of formaldehyde. Cell imaging in vitro. Nanoscale 2014, 6, 9071–9077. [Google Scholar] [CrossRef]
- Chen, J.; Li, Y.; Lv, K.; Zhong, W.; Wang, H.; Wu, Z.; Yi, P.; Jiang, J. Cyclam-functionalized carbon dots sensor for sensitive and selective detection of copper(II) ion and sulfide anion in aqueous media and its imaging in live cells. Sens. Actuators B Chem. 2016, 224, 298–306. [Google Scholar] [CrossRef]
- Yang, X.; Zhuo, Y.; Zhu, S.; Luo, Y.; Feng, Y.; Dou, Y. Novel and green synthesis of high-fluorescent carbon dots originated from honey for sensing and imaging. Biosens. Bioelectron. 2014, 60, 292–298. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, H.S. Green synthesis of luminescent nitrogen-doped carbon dots from milk and its imaging application. Anal. Chem. 2014, 86, 8902–8905. [Google Scholar] [CrossRef]
- Zhao, S.; Lan, M.; Zhu, X.; Xue, H.; Ng, T.W.; Meng, X.; Lee, C.S.; Wang, P.; Zhang, W. Green synthesis of bifunctional fluorescent carbon dots from garlic for cellular imaging and free radical scavenging. ACS Appl. Mater. Interfaces 2015, 7, 17054–17060. [Google Scholar] [CrossRef]
- Sahu, S.; Behera, B.; Maiti, T.K.; Mohapatra, S. Simple one-step synthesis of highly luminescent carbon dots from orange juice: Application as excellent bio-imaging agents. Chem. Commun. 2012, 48, 8835–8837. [Google Scholar] [CrossRef]
- Xue, M.; Zhan, Z.; Zou, M.; Zhang, L.; Zhao, S. Green synthesis of stable and biocompatible fluorescent carbon dots from peanut shells for multicolor living cell imaging. New J. Chem. 2016, 40, 1698–1703. [Google Scholar] [CrossRef]
- Yang, L.; Jiang, W.; Qiu, L.; Jiang, X.; Zuo, D.; Wang, D.; Yang, L. One pot synthesis of highly luminescent polyethylene glycol anchored carbon dots functionalized with a nuclear localization signal peptide for cell nucleus imaging. Nanoscale 2015, 7, 6104–6113. [Google Scholar] [CrossRef]
- Kang, Y.F.; Fang, Y.W.; Li, Y.H.; Li, W.; Yin, X.B. Nucleus-staining with biomolecule-mimicking nitrogen-doped carbon dots prepared by a fast neutralization heat strategy. Chem. Commun. 2015, 51, 16956–16959. [Google Scholar] [CrossRef]
- Datta, K.K.R.; Kozak, O.; Ranc, V.; Havrdova, M.; Bourlinos, A.B.; Safarova, K.; Hola, K.; Tomankova, K.; Zoppellaro, G.; Otyepka, M.; et al. Quaternized carbon dot-modified graphene oxide for selective cell labelling–controlled nucleus and cytoplasm imaging. Chem. Commun. 2014, 50, 10782–10785. [Google Scholar] [CrossRef]
- Kim, S.; Choi, Y.; Park, G.; Won, C.; Park, Y.J.; Lee, Y.; Kim, B.S.; Min, D.H. Highly efficient gene silencing and bioimaging based on fluorescent carbon dots in vitro and in vivo. Nano Res. 2017, 10, 503–519. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, X.; Fang, Z.; Niu, Y.; Lou, J.; Wu, Y.; Zou, S.; Xia, S.; Sun, M.; Du, F. Fabrication of HA/PEI-functionalized carbon dots for tumor targeting, intracellular imaging and gene delivery. RSC Adv. 2017, 7, 3369–3375. [Google Scholar] [CrossRef] [Green Version]
- Gao, G.; Jiang, Y.W.; Yang, J.J.; Wu, F.G. Mitochondria-targetable carbon quantum dots for differentiating cancerous cells from normal cells. Nanoscale 2017, 9, 18368–18378. [Google Scholar] [CrossRef]
- Hua, X.W.; Bao, Y.W.; Chen, Z.; Wu, F.G. Carbon quantum dots with intrinsic mitochondrial targeting ability for mitochondria-based theranostics. Nanoscale 2017, 9, 10948–10960. [Google Scholar] [CrossRef]
- Yuan, M.K.; Guo, Y.J.; Wei, J.J.; Li, J.Z.; Long, T.F.; Liu, Z.D. Optically active blue-emitting carbon dots to specifically target the Golgi apparatus. RSC Adv. 2017, 7, 49931–49936. [Google Scholar] [CrossRef] [Green Version]
- Li, R.S.; Gao, P.F.; Zhang, H.Z.; Zheng, L.L.; Li, C.M.; Wang, J.; Li, Y.F.; Liu, F.; Li, N.; Huang, C.Z. Chiral nanoprobes for targeting and long-term imaging of the Golgi apparatus. Chem. Sci. 2017, 8, 6829–6835. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Li, X.; Ling, Y.; Huang, C.; Jia, N. Morpholine derivate-functionalized carbon dots-based fluorescent probe for highly selective lysosomal imaging in living cells. ACS Appl. Mater. Interfaces 2017, 9, 28222–28232. [Google Scholar] [CrossRef]
- Raices, M.A.; D’Angelo, M. Nuclear pore complex composition: A new regulator of tissue-specific and developmental functions. Mol. Cell Biol. 2012, 13, 687–699. [Google Scholar] [CrossRef]
- Patel, S.S.; Belmont, J.B.; Sante, J.M.; Rexach, M.F. Natively unfolded nucloporins gate protein diffusion across the nuclear pore complex. Cell 2007, 129, 83–86. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, H.B.; Görlich, D. Transport selectivity of nuclear pores, phase separation, and membraneless organelles. Trends Biochem. Sci. 2016, 41, 46–61. [Google Scholar] [CrossRef]
- Samudram, A.; Mangalassery, B.M.; Kowshik, M.; Patincharath, N.; Varier, G.K. Passive permeability and effective pore size of HeLa cell nuclear membranes. Cell Biol. Int. 2016, 40, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Ori, A.; Banterle, N.; Iskar, M.; Andrés-Pons, A.; Escher, C.; Bui, H.K.; Sparks, L.; Solis-Meyarino, V.; Rinner, O.; Bork, P.; et al. Cell type-specific nuclear pores: A case in point for context-dependent stoichiometry of molecular machines. Mol. Syst. Biol. 2013, 9, 648. [Google Scholar] [CrossRef] [PubMed]
- Fahrenkrog, B.; Aebi, U. The nuclear pore complex: Nucleocytoplasmic transport and beyond. Nat. Rev. Mol. Cell Biol. 2003, 4, 757–766. [Google Scholar] [CrossRef] [PubMed]
- Stoffler, D.; Feja, B.; Fahrenkrog, B.; Walz, J.; Typke, D.; Aebi, U. Cryo-electron tomography provides novel insights into nuclear pore architecture—Implications for nucleocytoplasmic transport. J. Mol. Biol. 2003, 328, 119–130. [Google Scholar] [CrossRef]
- Stoffler, D.; Fahrenkrog, B.; Aebi, U. The nuclear pore complex: From molecular architecture to functional dynamics. Curr. Opin. Cell Biol. 1999, 11, 391–401. [Google Scholar] [CrossRef]
- Hinshaw, J.E.; Carragher, B.O.; Milligan, R.A. Architecture and design of the nuclear pore complex. Cell 1992, 69, 1133–1141. [Google Scholar] [CrossRef]
- Akey, C.W.; Radermacher, M. Architecture of the Xenopus nuclear pore complex revealed by 3-dimensional cryo-electron microscopy. J. Cell Biol. 1993, 122, 1–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Brattain, M.G. The maximal size of protein to diffuse through the nuclear pore is larger than 60 kDa. FEBS Lett. 2007, 581, 3164–3170. [Google Scholar] [CrossRef] [Green Version]
- Barua, S.; Mitragotri, S. Challenges associated with penetration of nanoparticles across cell and tissue barriers: A review of current status and future prospects. Nano Today 2014, 9, 223–243. [Google Scholar] [CrossRef]
- Tkachenko, A.G.; Xie, H.; Liu, Y.; Coleman, D.; Ryan, J.; Glomm, W.R.; Shipton, M.K.; Franzen, S.; Feldheim, D.L. Cellular trajectories of peptide-modified gold particle complex: Comparison of nuclear localization signals and peptide transduction domains. Bioconjug. Chem. 2004, 15, 482–490. [Google Scholar] [CrossRef]
- Havrdova, M.; Hola, K.; Skopalik, J.; Tomankova, K.; Petr, M.; Cepe, K.; Polakova, K.; Tucek, J.; Bourlinous, A.B.; Zboril, R. Toxicity of carbon dots—Effect of surface functionalization on the cell viability, reactive oxygen species generation and cell cycle. Carbon 2016, 99, 238–248. [Google Scholar] [CrossRef]
- Moghimi, S.; Symonds, P.; Murray, J.; Hunter, A.; Debska, G.; Szewczyk, A. A two-stage poly(ethylenimine)-mediated cytotoxicity: Implications for gene transfer/therapy. Mol. Ther. 2005, 11, 990–995. [Google Scholar] [CrossRef]
- Iida, T.; Mori, T.; Katayama, Y.; Niidome, T. Overall interaction of cytosolic proteins with the PEI/DNA complex. J. Control Release 2007, 18, 364–369. [Google Scholar] [CrossRef]
- Sahinturk, V.; Kacar, S.; Vejselova, D.; Kutlu, H.M. Acrylamide exerts its cytotoxicity in NIH/3T3 fibroblast cells by apoptosis. Toxicol. Ind. Health 2018, 34, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Cannella, V.; Altomare, R.; Chiaramonte, G.; Di Bella, S.; Mira, F.; Russotto, L.; Pisano, P.; Guercio, A. Cytotoxicity Evaluation of Endodontic Pins on L929 Cell Line. BioMed Res. Int. 2019, 3469525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozdemir, K.G.; Yilmaz, H.; Yilmaz, S. In vitro evaluation of cytotoxicity of soft lining materials on L929 cells by MTT assay. J. Biomed. Mater. Res. B Appl. Biomater. 2009, 90, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Scita, G.; Paolo, D.F.P. The endocytic matrix. Nature 2010, 28, 464–473. [Google Scholar] [CrossRef]
- Randow, F.; MacMicking, J.D.; James, L.C. Cellular self-defense: How cell-autonomous immunity protects against pathogens. Science 2013, 340, 701–706. [Google Scholar] [CrossRef]
- Yang, H.; Liu, Y.; Guo, Z.; Lei, B.; Zhuang, J.; Zhang, X.; Liu, Z.; Hu, C. Hydrophobic carbon dots with blue dispersed emission and red aggregation-induced emission. Nat. Commun. 2019, 10, 1789. [Google Scholar] [CrossRef]
- Arsov, Z.; Urbančič, I.; Štrancar, J. Aggregation-induced emission spectral shift as a measure of local concentration of a pH-activatable rhodamine-based smart probe. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2018, 190, 486–493. [Google Scholar] [CrossRef]
- Zhou, N.; Zhu, S.; Maharjan, S.; Hao, Z.; Song, Y.; Zhao, X.; Jiang, Y.; Yang, B.; Lu, L. Elucidating the endocytosis, intracellular trafficking, and exocytosis of carbon dots in neural cells. RSC Adv. 2014, 4, 62086–62095. [Google Scholar] [CrossRef]
- Francia, V.; Reker-Smit, C.; Boel, G.; Slavati, A. Limits and challenges in using transport inhibitors to characterize how nano-sized drug carriers enter cells. Nanomedicine 2019, 14, 1533–1549. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, P.A.A.; Boucrot, E. Mechanisms of carrier formation during clathrin-independent endocytosis. Trends Cell Biol. 2018, 28, 188–200. [Google Scholar] [CrossRef]
- Rejman, J.; Oberle, V.; Zuhorn, I.S.; Hoekstra, D. Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis. Biochem. J. 2004, 377, 159–169. [Google Scholar] [CrossRef]
- Vercauteren, D.; Vandenbroucke, R.E.; Jones, A.T.; Rejman, J.; Demeester, J.; De Smedt, S.C.; Sanders, N.N.; Braeckmans, K. The use of inhibitors to study endocytic pathways of gene carriers: Optimization and pitfalls. Mol. Ther. 2010, 18, 561–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.H.; Rothberg, K.G.; Anderson, R.G. Mis-assemblz of clatrin lattice on endosomes reveals a regulatory switch for coated pit formation. J. Cell Biol. 1993, 123, 1107–1117. [Google Scholar] [CrossRef]
- Chen, F.; Zhu, L.; Zhang, Y.; Kumar, D.; Cao, G.; Hu, X.; Liang, Z.; Kuang, S.; Xue, R.; Gong, C. Clathrin-mediated endocytosis is a candidate entry sorting mechanism for Bombyx mori cypovirus. Sci. Rep. 2018, 8, 7268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subtil, A.; Dautry-Varsat, A. Microtubule depolymerization inhibits clatrin coated-pit internalization in non-adherent cell lines while interleukin 2 endocytosis is not affected. J. Cell Sci. 1997, 110, 2441–2447. [Google Scholar] [CrossRef] [PubMed]
- Mundz, D.I.; Machleidt, T.; Ying, Y.S.; Anderson, R.G.; Bloom, G.S. Dual control of caveolar membrane traffic by microtubules and the actin cytoskeleton. J. Cell Sci. 2002, 115, 4327–4339. [Google Scholar] [CrossRef] [Green Version]
- Caviston, J.P.; Holzbaur, E.L. Microtubule motors at the intersection of trafficking and transport. Trends Cell Biol. 2006, 16, 530–537. [Google Scholar] [CrossRef]
- Jordan, M.A.; Thrower, D.; Wilson, L. Effects of vinblastine, podophyllotoxin and nocodazole on mitotic spindles. Implications for the role of microtubule dynamics in mitosis. J. Cell Sci. 1992, 102, 401–416. [Google Scholar] [CrossRef]
- Dos Santos, T.; Varela, J.; Lynch, I.; Salvati, A.; Dawson, K.A. Effects of transport inhibitors on the cellular uptake of carboxylated polystyrene nanoparticles in different cell lines. PLoS ONE 2011, 6, e24438. [Google Scholar] [CrossRef] [Green Version]
- Rémy-Kristensen, A.; Clamme, J.P.; Vuilleumier, C.; Kuhry, J.G.; Mély, Y. Role of endocytosis in the transfection of L929 fibroblasts by polyethylenimine/DNA complexes. Biochim. Biophys. Acta Biomembr. 2001, 1514, 21–32. [Google Scholar] [CrossRef] [Green Version]
- Chithrani Devika, B.; Chan, C.W.W. Elucidating the Mechanism of Cellular Uptake and Removal of Protein-Coated Gold Nanoparticles of Different Sizes and Shapes. Nano Lett. 2007, 7, 1542–1550. [Google Scholar] [CrossRef]
- Foster, I. Cancer: A cell cycle defect. Radiography 2008, 14, 144–149. [Google Scholar] [CrossRef]
- Bourlinos, A.B.; Zboril, R.; Petr, J.; Bakandritsos, A.; Krysmann, M.; Giannelis, E.P. Luminescent surface quaternized carbon dots. Chem. Mater. 2012, 24, 6–8. [Google Scholar] [CrossRef]
- Malina, T.; Polakova, K.; Skopalik, J.; Milotova, V.; Hola, K.; Havrdova, M.; Tomankova, K.B.; Cmiel, V.; Sefc, L.; Zboril, R. Carbon dots for in vivo fluorescence imaging of adipose tissue-derived mesenchymal stromal cells. Carbon 2019, 152, 434–443. [Google Scholar] [CrossRef]
- Krysmann, M.J.; Kelarakis, A.; Dallas, P.; Giannelis, E.P. Formation mechanism of carbogenic nanoparticles with dual photoluminescence emission. J. Am. Chem. Soc. 2012, 134, 747–750. [Google Scholar] [CrossRef]
- Essner, J.B.; Kist, J.A.; Polo-Parada, L.; Baker, G.A. Artifacts and errors associated with the ubiquitous presence of fluorescent impurities in carbon nanodots. Chem. Mater. 2018, 30, 1878–1887. [Google Scholar] [CrossRef]
- Arsov, Z.; Urbančič, I.; Garvas, M.; Biglino, D.; Ljubetič, A.; Koklič, T.; Štrancar, J. Fluorescence microspectroscopy as a tool to study mechanism of nanoparticles delivery into living cancer cells. Biomed. Opt. Express 2011, 2, 2083–2095. [Google Scholar] [CrossRef] [Green Version]
- Urbančič, I.; Arsov, Z.; Ljubetič, A.; Biglino, D.; Štrancar, J. Bleaching-corrected fluorescence microspectroscopy with nanometer peak position resolution. Opt. Express 2013, 21, 25291–25306. [Google Scholar] [CrossRef]
- Wang, Y.; Kalytchuk, S.; Zhang, Y.; Shi, H.; Kershaw, S.V.; Rogach, A.L. Thickness dependent full-color emission tunability in a flexible carbon dot ionogel. J. Phys. Chem. Lett. 2014, 5, 1412–1420. [Google Scholar] [CrossRef]
- Pan, L.; Sun, S.; Zhang, A.; Jiang, K.; Zhang, L.; Dong, C.; Huang, Q.; Wu, A.; Lin, H. Truly Fluorescent Excitation-Dependent Carbon Dots and Their Applications in Multicolor. Cellular Imaging and Multidimensional Sensing. Adv. Mater. 2015, 27, 7782–7787. [Google Scholar] [CrossRef]
- Collins, A.R. The comet assay for DNA damage and repair: Principles, applications, and limitations. Mol. Biotechnol. 2004, 26, 249–261. [Google Scholar] [CrossRef]
- Tomankova, K.; Kejlova, K.; Binder, S.; Daskova, A.; Zapletalová, J.; Bendová, H.; Kolarova, H.; Jirova, D. In vitro cytotoxicity and phototoxicity study of cosmetics colorants. Toxicol. In Vitro 2011, 25, 1242–1250. [Google Scholar] [CrossRef]










Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Havrdová, M.; Urbančič, I.; Bartoň Tománková, K.; Malina, L.; Štrancar, J.; Bourlinos, A.B. Self-Targeting of Carbon Dots into the Cell Nucleus: Diverse Mechanisms of Toxicity in NIH/3T3 and L929 Cells. Int. J. Mol. Sci. 2021, 22, 5608. https://doi.org/10.3390/ijms22115608
Havrdová M, Urbančič I, Bartoň Tománková K, Malina L, Štrancar J, Bourlinos AB. Self-Targeting of Carbon Dots into the Cell Nucleus: Diverse Mechanisms of Toxicity in NIH/3T3 and L929 Cells. International Journal of Molecular Sciences. 2021; 22(11):5608. https://doi.org/10.3390/ijms22115608
Chicago/Turabian StyleHavrdová, Markéta, Iztok Urbančič, Kateřina Bartoň Tománková, Lukáš Malina, Janez Štrancar, and Athanasios B. Bourlinos. 2021. "Self-Targeting of Carbon Dots into the Cell Nucleus: Diverse Mechanisms of Toxicity in NIH/3T3 and L929 Cells" International Journal of Molecular Sciences 22, no. 11: 5608. https://doi.org/10.3390/ijms22115608
APA StyleHavrdová, M., Urbančič, I., Bartoň Tománková, K., Malina, L., Štrancar, J., & Bourlinos, A. B. (2021). Self-Targeting of Carbon Dots into the Cell Nucleus: Diverse Mechanisms of Toxicity in NIH/3T3 and L929 Cells. International Journal of Molecular Sciences, 22(11), 5608. https://doi.org/10.3390/ijms22115608