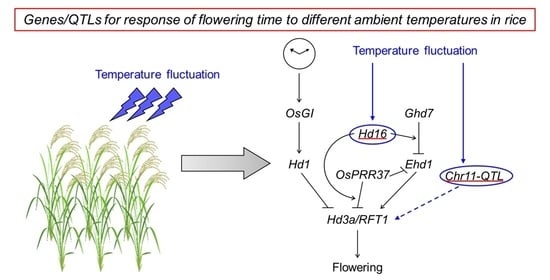
Genetic Elucidation for Response of Flowering Time to Ambient Temperatures in Asian Rice Cultivars
Abstract
:1. Introduction
2. Results
2.1. Natural Variations in Flowering Time Among Years
2.2. Flowering Time Fluctuation in CSSLs of “Koshihikari” and “Khao Nam Jen”
2.3. Transcriptome Analysis in Rice Plants from Juvenile to Mature Stages
2.4. Effect of Weather Factors on Flowering Time in Rice Plants
3. Discussion
3.1. Detection of QTLs for Response of Flowering Time to Ambient Temperature Fluctuations
3.2. Integration of Thermal Response into Photoperiod Flowering Pathway
3.3. Application to Development of Climate-Resilient Crops
4. Conclusions
5. Materials and Methods
5.1. Plant Materials
5.2. Evaluation of Flowering Time in Natural Field Conditions
5.3. QTL Detection in CSSLs and NILs
5.4. Transcriptome at Panicle Initiation Stage
5.5. Correlation between Flowering Time and Meteorological Conditions
5.6. Evaluation of Flowering Time under High and Low Temperatures
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- IPCC (Intergovernmental Panel on Climate Change). Global Warming of 1.5 °C. 2018. Available online: https://report.ipcc.ch/sr15/pdf/sr15_spm_final.pdf (accessed on 19 January 2021).
- Londo, J.P.; Chiang, Y.C.; Hung, K.H.; Chiang, T.Y.; Schaal, B.A. Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc. Natl. Acad. Sci. USA 2006, 103, 9578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Kurata, N.; Wei, X.; Wang, Z.X.; Wang, A.; Zhao, Q.; Zhao, Y.; Liu, K.; Lu, H.; Li, W.; et al. A map of rice genome variation reveals the origin of cultivated rice. Nature 2012, 490, 497–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bathiany, S.; Dakos, V.; Scheffer, M.; Lenton, T.M. Climate models predict increasing temperature variability in poor countries. Sci. Adv. 2018, 4, eaar5809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeuchi, Y.; Hayasaka, H.; Chiba, B.; Tanaka, I.; Shimano, T.; Yamagishi, M.; Nagano, K.; Sasaki, T.; Yano, M. Mapping quantitative trait loci controlling cool-temperature tolerance at booting stage in temperate japonica rice. Breed. Sci. 2001, 51, 191–197. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; Yang, C.; Xu, Q.; Wang, L.; Yang, X.; Song, X.; Wang, J.; Zhang, X.; Li, B.; Li, H.; et al. Genetic dissection of germinability under low temperature by building a resequencing linkage map in japonica rice. Int. J. Mol. Sci. 2020, 21, 1284. [Google Scholar] [CrossRef] [Green Version]
- Thomas, B.; Vince-Prue, D. Photoperiodism in Plants, 2nd ed.; Academic Press: San Diego, CA, USA, 1997. [Google Scholar]
- Izawa, T. Daylength measurements by rice plants in photoperiodic short-day flowering. In International Review of Cytology; Kwang, W.J., Ed.; Academic Press: Cambridge, MA, USA, 2007; Volume 256, pp. 191–222. [Google Scholar]
- Shrestha, R.; Gomez-Ariza, J.; Brambilla, V.; Fornara, F. Molecular control of seasonal flowering in rice, Arabidopsis and temperate cereals. Ann. Bot. 2014, 114, 1445–1458. [Google Scholar] [CrossRef] [Green Version]
- Hori, K.; Matsubara, K.; Yano, M. Genetic control of flowering time in rice: Integration of Mendelian genetics and genomics. Theor. Appl. Genet. 2016, 129, 2241–2252. [Google Scholar] [CrossRef]
- Brambilla, V.; Martignago, D.; Goretti, D.; Cerise, M.; Somssich, M.; De Rosa, M.; Galbiati, F.; Simon, R.; Lazzaro, F.; Simon, R.; et al. Antagonistic transcription factor complexes modulate the floral transition in rice. Plant Cell 2017, 29, 2801. [Google Scholar] [CrossRef] [Green Version]
- Matsubara, K.; Yano, M. Genetic and molecular dissection of flowering time control in rice. In Rice Genomics, Genetics and Breeding; Sasaki, T., Ashikari, M., Eds.; Springer: Singapore, 2018; pp. 177–190. [Google Scholar]
- Liu, H.; Zhou, X.; Li, Q.; Wang, L.; Xing, Y. CCT domain-containing genes in cereal crops: Flowering time and beyond. Theor. Appl. Genet. 2020, 133, 1385–1396. [Google Scholar] [CrossRef]
- Qüesta, J.I.; Song, J.; Geraldo, N.; An, H.; Dean, C. Arabidopsis transcriptional repressor VAL1 triggers Polycomb silencing at FLC during vernalization. Science 2016, 353, 485–488. [Google Scholar] [CrossRef]
- Bloomer, R.H.; Dean, C. Fine-tuning timing: Natural variation informs the mechanistic basis of the switch to flowering in Arabidopsis thaliana. J. Exp. Bot. 2017, 68, 5439–5452. [Google Scholar] [CrossRef]
- Jung, J.H.; Domijan, M.; Klose, C.; Biswas, S.; Ezer, D.; Gao, M.; Khattak, A.K.; Box, M.S.; Charoensawan, V.; Cortijo, S.; et al. Phytochromes function as thermosensors in Arabidopsis. Science 2016, 354, 886–889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Hwang, G.; Kim, S.; Thi, T.N.; Kim, H.; Jeong, J.; Kim, J.; Kim, J.; Choi, G.; Oh, E. The epidermis coordinates thermoresponsive growth through the phyB-PIF4-auxin pathway. Nat. Commun. 2020, 11, 1053. [Google Scholar] [CrossRef] [Green Version]
- Hahm, J.; Kim, K.; Qiu, Y.; Chen, M. Increasing ambient temperature progressively disassembles Arabidopsis phytochrome B from individual photobodies with distinct thermostabilities. Nat. Commun. 2020, 11, 1660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanco-Touriñán, N.; Legris, M.; Minguet, E.G.; Costigliolo-Rojas, C.; Nohales, M.A.; Iniesto, E.; García-León, M.; Pacín, M.; Heucken, N.; Blomeier, T.; et al. COP1 destabilizes DELLA proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 2020, 117, 13792–13799. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Barbosa, A.D.; Hutin, S.; Kumita, J.R.; Gao, M.; Derwort, D.; Silva, C.S.; Lai, X.; Pierre, E.; Geng, F.; et al. A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature 2020, 585, 256–260. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C. Effect of seasonal changes of day-length and temperature to crop growth I. Studies on growth period of rice cultivars in Taiwan. Agric. Hortic. 1938, 13, 21–32. [Google Scholar]
- Wada, E. Studies on the response of heading to day-length and temperature in rice plants. I. Response of varieties and the relation to their geographical distribution in Japan. Jpn. J. Breed. 1952, 2, 55–62. [Google Scholar] [CrossRef]
- Oka, H. Varietal variation of the responses to day-length and temperature and the number of days to growth period. Phylogenic differentiation of the cultivated rice plant. III. Japan. Jpn. J. Breed. 1954, 4, 92–100. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, H.; Yamagishi, J.; Miyamoto, N.; Motoyama, M.; Yano, M.; Nemoto, K. Flowering response of rice to photoperiod and temperature: A QTL analysis using a phenological model. Theor. Appl. Genet. 2005, 110, 778–786. [Google Scholar] [CrossRef]
- Luan, W.; Chen, H.; Fu, Y.; Si, H.; Peng, W.; Song, S.; Liu, W.; Hu, G.; Sun, Z.; Xie, D.; et al. The effect of the crosstalk between photoperiod and temperature on the heading date in rice. PLoS ONE 2009, 4, e5891. [Google Scholar] [CrossRef]
- Song, Y.; Gao, Z.; Luan, W. Interaction between temperature and photoperiod in regulation of flowering time in rice. Sci. China Life Sci. 2012, 55, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Nagata, K.; Nonoue, Y.; Mizobuchi, R.; Ono, N.; Shibaya, T.; Ebana, K.; Matsubara, K.; Ogiso, E. Development of twelve sets of chromosomal segment substitution lines that enhance allele mining in cultivated rice. Breed. Res. 2015, 17, 131. [Google Scholar]
- Takahashi, Y.; Shomura, A.; Sasaki, T.; Yano, M. Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the alpha subunit of protein kinase CK2. Proc. Natl. Acad. Sci. USA 2001, 98, 7922–7927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hori, K.; Ogiso-Tanaka, E.; Matsubara, K.; Yamanouchi, U.; Ebana, K.; Yano, M. Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response. Plant J. 2013, 76, 36–46. [Google Scholar] [CrossRef] [Green Version]
- Hori, K.; Nonoue, Y.; Ono, N.; Shibaya, T.; Ebana, K.; Matsubara, K.; Ogiso-Tanaka, E.; Tanabata, T.; Sugimoto, K.; Taguchi-Shiobara, F.; et al. Genetic architecture of variation in heading date among Asian rice accessions. BMC Plant Biol. 2015, 15, 115. [Google Scholar] [CrossRef] [Green Version]
- Itoh, H.; Wada, K.C.; Sakai, H.; Shibasaki, K.; Fukuoka, S.; Wu, J.; Yonemaru, J.-I.; Yano, M.; Izawa, T. Genomic adaptation of flowering-time genes during the expansion of rice cultivation area. Plant J. 2018, 94, 895–909. [Google Scholar] [CrossRef]
- Dai, C.; Xue, H.W. Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J. 2010, 29, 1916–1927. [Google Scholar] [CrossRef] [Green Version]
- Kwon, C.T.; Yoo, S.C.; Koo, B.H.; Cho, S.H.; Park, J.W.; Zhang, Z.; Li, J.; Li, Z.; Paek, N.C. Natural variation in Early flowering1 contributes to early flowering in japonica rice under long days. Plant Cell Environ. 2014, 37, 101–112. [Google Scholar] [CrossRef]
- Fornara, F.; de Montaigu, A.; Coupland, G. SnapShot: Control of flowering in Arabidopsis. Cell 2010, 141, 550–550.e2. [Google Scholar] [CrossRef] [Green Version]
- Leijten, W.; Koes, R.; Roobeek, I.; Frugis, G. Translating flowering time from Arabidopsis thaliana to Brassicaceae and Asteraceae crop species. Plants 2018, 7, e111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bentley, A.; Jensen, E.; Mackay, I.; Honicka, H.; Fladung, M.; Hori, K.; Yano, M.; Mullet, J. Flowering Time: Genomics and Breeding for Climate-Resilient Crops; Springer: Berlin, Germany, 2013; Volume 2, pp. 1–66. [Google Scholar]
- Henderson, I.R.; Shindo, C.; Dean, C. The need for winter in the switch to flowering. Annu. Rev. Genet. 2003, 37, 371–392. [Google Scholar] [CrossRef] [PubMed]
- Trevaskis, B.; Hemming, M.N.; Dennis, E.S.; Peacock, W.J. The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 2007, 12, 352–357. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Chong, K. Remembering winter through vernalisation. Nat. Plants 2018, 4, 997–1009. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Lin, H.; Chen, A.; Lau, M.; Jernstedt, J.; Dubcovsky, J. Wheat VRN1, FUL2 and FUL3 play critical and redundant roles in spikelet development and spike determinacy. Development 2019, 146, dev175398. [Google Scholar] [CrossRef] [Green Version]
- Woods, D.P.; McKeown, M.A.; Dong, Y.; Preston, J.C.; Amasino, R.M. Evolution of VRN2/Ghd7-like genes in vernalization-mediated repression of grass flowering. Plant Physiol. 2016, 170, 2124. [Google Scholar] [CrossRef] [Green Version]
- Hori, K.; Shenton, M. Recent advances in molecular research in rice: Agronomically important traits. Int. J. Mol. Sci. 2020, 21, 5945. [Google Scholar] [CrossRef]
- Fukuoka, S.; Nonoue, Y.; Yano, M. Germplasm enhancement by developing advanced plant materials from diverse rice accessions. Breed. Sci. 2010, 60, 509–517. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, Y. Developing isogenic lines of Japanese rice cultivar “Koshihikari” with early and late heading. JARQ 2007, 45, 15–22. [Google Scholar] [CrossRef] [Green Version]
- Sakai, H.; Lee, S.S.; Tanaka, T.; Numa, H.; Kim, J.; Kawahara, Y.; Wakimoto, H.; Yang, C.; Iwamoto, M.; Abe, T.; et al. Rice annotation project database (RAP-DB): An integrative and interactive database for rice genomics. Plant Cell Physiol. 2013, 54, e6. [Google Scholar] [CrossRef]
- Liu, L.; Mei, Q.; Yu, Z.; Sun, T.; Zhang, Z.; Chen, M. An integrative bioinformatics framework for genome-scale multiple level network reconstruction of rice. J. Integr. Bioinform. 2013, 10, 94–102. [Google Scholar] [CrossRef]
- Hayashi, Y.; Toritani, H.; Goto, S.; Yokozawa, M.; Seino, H. Climatic records of the National Institute of Agro-Environmental Sciences (1990–1996). Misc. Publ. Natl. Inst. Agro-Environ. Sci. 1998, 23, 1–168. [Google Scholar]





| Days to Heading | ||||||
|---|---|---|---|---|---|---|
| ‘Koshihikari’ | ‘Khao Nam Jen’ | SL2812 | SL2833 | Koshihikari Hd6-NIL | Koshihikari Hd16-NIL | |
| Maximum temperature | ||||||
| June | −0.07 | 0.34 *** | −0.08 | −0.01 | 0.14 | 0.48 *** |
| July | −0.26 *** | −0.20 | −0.35 *** | −0.45 *** | −0.34 *** | −0.44 *** |
| August | −0.16 | −0.32 *** | −0.18 | −0.24 | −0.24 | −0.42 *** |
| September | 0.03 | −0.43 *** | 0.20 | 0.16 | −0.02 | −0.63 *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hori, K.; Saisho, D.; Nagata, K.; Nonoue, Y.; Uehara-Yamaguchi, Y.; Kanatani, A.; Shu, K.; Hirayama, T.; Yonemaru, J.-i.; Fukuoka, S.; et al. Genetic Elucidation for Response of Flowering Time to Ambient Temperatures in Asian Rice Cultivars. Int. J. Mol. Sci. 2021, 22, 1024. https://doi.org/10.3390/ijms22031024
Hori K, Saisho D, Nagata K, Nonoue Y, Uehara-Yamaguchi Y, Kanatani A, Shu K, Hirayama T, Yonemaru J-i, Fukuoka S, et al. Genetic Elucidation for Response of Flowering Time to Ambient Temperatures in Asian Rice Cultivars. International Journal of Molecular Sciences. 2021; 22(3):1024. https://doi.org/10.3390/ijms22031024
Chicago/Turabian StyleHori, Kiyosumi, Daisuke Saisho, Kazufumi Nagata, Yasunori Nonoue, Yukiko Uehara-Yamaguchi, Asaka Kanatani, Koka Shu, Takashi Hirayama, Jun-ichi Yonemaru, Shuichi Fukuoka, and et al. 2021. "Genetic Elucidation for Response of Flowering Time to Ambient Temperatures in Asian Rice Cultivars" International Journal of Molecular Sciences 22, no. 3: 1024. https://doi.org/10.3390/ijms22031024
APA StyleHori, K., Saisho, D., Nagata, K., Nonoue, Y., Uehara-Yamaguchi, Y., Kanatani, A., Shu, K., Hirayama, T., Yonemaru, J. -i., Fukuoka, S., & Mochida, K. (2021). Genetic Elucidation for Response of Flowering Time to Ambient Temperatures in Asian Rice Cultivars. International Journal of Molecular Sciences, 22(3), 1024. https://doi.org/10.3390/ijms22031024