The Vps13 Family of Lipid Transporters and Its Role at Membrane Contact Sites
Abstract
:1. Introduction
2. Vps13 Belongs to a New Family of Lipid Transport Proteins
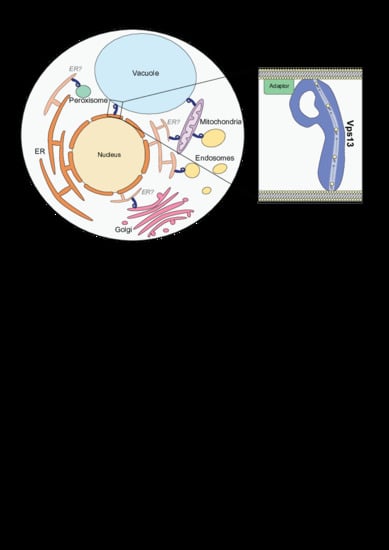
3. Vps13 Has a Variety of Localization Determinants
3.1. The N-Terminus as an ER-Targeting Determinant
3.2. Organelle-Specific Adaptors Target Vps13 to Membranes via a Conserved Motif
3.3. Vps13 Adaptor Binding Domain
3.4. Other Localization Determinants at the C-Terminus
3.4.1. APT1 Domain
3.4.2. ATG2_C Domains and Conserved Amphipathic Helices
3.4.3. PH Domain
4. Function of Vps13 at Specific Contact Sites
4.1. Vps13 at the NVJ
4.2. Vps13 at Mitochondrial Contact Sites
4.3. Vps13 and Prospore Membrane Expansion
4.4. VPS13 at Autophagosomal Membranes
4.5. VPS13 Proteins at LDs
4.6. Vps13 at Other Contact Sites
5. Drivers of Lipid Transport
5.1. The Role of Lipid Biosynthesis
5.2. The Role of Lipid Scramblases
6. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Jain, A.; Holthuis, J.C.M. Membrane contact sites, ancient and central hubs of cellular lipid logistics. Biochim. Biophys. Acta 2017, 1864, 1450–1458. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.H.; Gatta, A.T.; Levine, T.P. Lipid transfer proteins: The lipid commute via shuttles, bridges and tubes. Nat. Rev. Mol. Cell Biol. 2019, 20, 85–101. [Google Scholar] [CrossRef]
- Nishimura, A.L.; Mitne-Neto, M.; Silva, H.C.A.; Richieri-Costa, A.; Middleton, S.; Cascio, D.; Kok, F.; Oliveira, J.R.M.; Gillingwater, T.; Webb, J.; et al. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am. J. Hum. Genet. 2004, 75, 822–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loewen, C.J.R.; Levine, T.P. A highly conserved binding site in vesicle-associated membrane protein-associated protein (VAP) for the FFAT motif of lipid-binding proteins. J. Biol. Chem. 2005, 280, 14097–14104. [Google Scholar] [CrossRef] [Green Version]
- Bryant, D.; Liu, Y.; Datta, S.; Hariri, H.; Seda, M.; Anderson, G.; Peskett, E.; Demetriou, C.; Sousa, S.; Jenkins, D.; et al. SNX14 mutations affect endoplasmic reticulum-associated neutral lipid metabolism in autosomal recessive spinocerebellar ataxia 20. Hum. Mol. Genet. 2018, 27, 1927–1940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Datta, S.; Liu, Y.; Hariri, H.; Bowerman, J.; Henne, W.M. Cerebellar ataxia disease-associated Snx14 promotes lipid droplet growth at ER-droplet contacts. J. Cell Biol. 2019, 218, 1335–1351. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Wang, J.; Coutavas, E.; Shi, H.; Hao, Q.; Blobel, G. Structure of human Niemann-Pick C1 protein. Proc. Natl. Acad. Sci. USA 2016, 113, 8212–8217. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.L.; Motamed, M.; Infante, R.E.; Abi-Mosleh, L.; Kwon, H.J.; Brown, M.S.; Goldstein, J.L. Identification of surface residues on Niemann-Pick C2 essential for hydrophobic handoff of cholesterol to NPC1 in lysosomes. Cell Metab. 2010, 12, 166–173. [Google Scholar] [CrossRef] [Green Version]
- Rampoldi, L.; Dobson-Stone, C.; Rubio, J.P.; Danek, A.; Chalmers, R.M.; Wood, N.W.; Verellen, C.; Ferrer, X.; Malandrini, A.; Fabrizi, G.M.; et al. A conserved sorting-associated protein is mutant in chorea-acanthocytosis. Nat. Genet. 2001, 28, 119–120. [Google Scholar] [CrossRef] [PubMed]
- Kolehmainen, J.; Black, G.C.M.; Saarinen, A.; Chandler, K.; Clayton-Smith, J.; Träskelin, A.-L.; Perveen, R.; Kivitie-Kallio, S.; Norio, R.; Warburg, M.; et al. Cohen syndrome is caused by mutations in a novel gene, COH1, encoding a transmembrane protein with a presumed role in vesicle-mediated sorting and intracellular protein transport. Am. J. Hum. Genet. 2003, 72, 1359–1369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lesage, S.; Drouet, V.; Majounie, E.; Deramecourt, V.; Jacoupy, M.; Nicolas, A.; Cormier-Dequaire, F.; Hassoun, S.M.; Pujol, C.; Ciura, S.; et al. Loss of VPS13C function in autosomal-recessive parkinsonism causes mitochondrial dysfunction and increases PINK1 / Parkin-dependent mitophagy. Am. J. Hum. Genet. 2016, 98, 500–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seong, E.; Insolera, R.; Dulovic, M.; Kamsteeg, E.-J.; Trinh, J.; Brüggemann, N.; Sandford, E.; Li, S.; Ozel, A.B.; Li, J.Z.; et al. Mutations in VPS13D lead to a new recessive ataxia with spasticity and mitochondrial defects. Ann. Neurol. 2018, 83, 1075–1088. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, J.; Meijer, I.A.; Lessel, D.; Mencacci, N.E.; Krainc, D.; Hempel, M.; Tsiakas, K.; Prokisch, H.; Rossignol, E.; Helm, M.H.; et al. Recessive mutations in >VPS13D cause childhood onset movement disorders. Ann. Neurol. 2018, 83, 1089–1095. [Google Scholar] [CrossRef] [PubMed]
- Seifert, W.; Kühnisch, J.; Maritzen, T.; Horn, D.; Haucke, V.; Hennies, H.C. Cohen syndrome-associated protein, COH1, is a novel, giant Golgi matrix protein required for Golgi integrity. J. Biol. Chem. 2011, 286, 37665–37675. [Google Scholar] [CrossRef] [Green Version]
- Kumar, N.; Leonzino, M.; Hancock-Cerutti, W.; Horenkamp, F.A.; Li, P.; Lees, J.A.; Wheeler, H.; Reinisch, K.M.; De Camilli, P. VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 2018, 217, 3625–3639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeshaw, W.M.; van der Zwaag, M.; Pinto, F.; Lahaye, L.L.; Faber, A.I.; Gómez-Sánchez, R.; Dolga, A.M.; Poland, C.; Monaco, A.P.; van IJzendoorn, S.C.; et al. Human VPS13A is associated with multiple organelles and influences mitochondrial morphology and lipid droplet motility. Elife 2019, 8. [Google Scholar] [CrossRef]
- Koike, S.; Jahn, R. SNAREs define targeting specificity of trafficking vesicles by combinatorial interaction with tethering factors. Nat. Commun. 2019, 10, 1608. [Google Scholar] [CrossRef] [PubMed]
- Bean, B.D.M.; Dziurdzik, S.K.; Kolehmainen, K.L.; Fowler, C.M.S.; Kwong, W.K.; Grad, L.I.; Davey, M.; Schluter, C.; Conibear, E. Competitive organelle-specific adaptors recruit Vps13 to membrane contact sites. J. Cell Biol. 2018, 217, 3593–3607. [Google Scholar] [CrossRef] [Green Version]
- Ugur, B.; Hancock-Cerutti, W.; Leonzino, M.; De Camilli, P. Role of VPS13, a protein with similarity to ATG2, in physiology and disease. Curr. Opin. Genet. Dev. 2020, 65, 61–68. [Google Scholar] [CrossRef]
- Muñoz-Braceras, S.; Tornero-Écija, A.R.; Vincent, O.; Escalante, R. VPS13A is closely associated with mitochondria and is required for efficient lysosomal degradation. Dis. Model. Mech. 2019, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seifert, W.; Holder-Espinasse, M.; Spranger, S.; Hoeltzenbein, M.; Rossier, E.; Dollfus, H.; Lacombe, D.; Verloes, A.; Chrzanowska, K.H.; Maegawa, G.H.B.; et al. Mutational spectrum of COH1 and clinical heterogeneity in Cohen syndrome. J. Med. Genet. 2006, 43, e22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Costa, R.; Bordessoules, M.; Guilleman, M.; Carmignac, V.; Lhussiez, V.; Courot, H.; Bataille, A.; Chlémaire, A.; Bruno, C.; Fauque, P.; et al. Vps13b is required for acrosome biogenesis through functions in Golgi dynamic and membrane trafficking. Cell. Mol. Life Sci. 2020, 77, 511–529. [Google Scholar] [CrossRef] [PubMed]
- Rogers, L.D.; Foster, L.J. The dynamic phagosomal proteome and the contribution of the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2007, 104, 18520–18525. [Google Scholar] [CrossRef] [Green Version]
- Shui, W.; Sheu, L.; Liu, J.; Smart, B.; Petzold, C.J.; Hsieh, T.Y.; Pitcher, A.; Keasling, J.D.; Bertozzi, C.R. Membrane proteomics of phagosomes suggests a connection to autophagy. Proc. Natl. Acad. Sci. USA 2008, 105, 16952–16957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trost, M.; English, L.; Lemieux, S.; Courcelles, M.; Desjardins, M.; Thibault, P. The Phagosomal Proteome in Interferon-γ-Activated Macrophages. Immunity 2009, 30, 143–154. [Google Scholar] [CrossRef] [Green Version]
- Guillén-Samander, A.; Leonzino, M.; Hanna, M.G., IV; Tang, N.; Shen, H.; De Camilli, P. VPS13D bridges the ER to Miro containing membranes. bioRxiv 2020. [Google Scholar] [CrossRef]
- Lang, A.B.; John Peter, A.T.; Walter, P.; Kornmann, B. ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. J. Cell Biol. 2015, 210, 883–890. [Google Scholar] [CrossRef] [Green Version]
- Park, J.-S.; Thorsness, M.K.; Policastro, R.; McGoldrick, L.L.; Hollingsworth, N.M.; Thorsness, P.E.; Neiman, A.M. Yeast Vps13 promotes mitochondrial function and is localized at membrane contact sites. Mol. Biol. Cell 2016, 27, 2435–2449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.-S.; Neiman, A.M. VPS13 regulates membrane morphogenesis during sporulation in Saccharomyces cerevisiae. J. Cell Sci. 2012, 125, 3004–3011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rzepnikowska, W.; Flis, K.; Kaminska, J.; Grynberg, M.; Urbanek, A.; Ayscough, K.R.; Zoladek, T. Amino acid substitution equivalent to human chorea-acanthocytosis I2771R in yeast Vps13 protein affects its binding to phosphatidylinositol 3-phosphate. Hum. Mol. Genet. 2017, 26, 1497–1510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- John Peter, A.T.; Herrmann, B.; Antunes, D.; Rapaport, D.; Dimmer, K.S.; Kornmann, B. Vps13-Mcp1 interact at vacuole-mitochondria interfaces and bypass ER-mitochondria contact sites. J. Cell Biol. 2017, 216, 3219–3229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Mari, M.; Parashar, S.; Liu, D.; Cui, Y.; Reggiori, F.; Novick, P.J.; Ferro-Novick, S. Vps13 is required for the packaging of the ER into autophagosomes during ER-phagy. Proc. Natl. Acad. Sci. USA 2020, 117, 18530–18539. [Google Scholar] [CrossRef] [PubMed]
- De, M.; Oleskie, A.N.; Ayyash, M.; Dutta, S.; Mancour, L.; Abazeed, M.E.; Brace, E.J.; Skiniotis, G.; Fuller, R.S. The Vps13p-Cdc31p complex is directly required for TGN late endosome transport and TGN homotypic fusion. J. Cell Biol. 2017, 216, 425–439. [Google Scholar] [CrossRef]
- Bankaitis, V.A.; Johnson, L.M.; Emr, S.D. Isolation of yeast mutants defective in protein targeting to the vacuole. Proc. Natl. Acad. Sci. USA 1986, 83, 9075–9079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kornmann, B.; Currie, E.; Collins, S.R.; Schuldiner, M.; Nunnari, J.; Weissman, J.S.; Walter, P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 2009, 325, 477–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopec, K.O.; Alva, V.; Lupas, A.N. Homology of SMP domains to the TULIP superfamily of lipid-binding proteins provides a structural basis for lipid exchange between ER and mitochondria. Bioinformatics 2010, 26, 1927–1931. [Google Scholar] [CrossRef] [Green Version]
- Li, P.Q.; Lees, J.A.; Lusk, C.P.; Reinisch, K.M. Cryo-EM reconstruction of a VPS13 fragment reveals a long groove to channel lipids between membranes. J. Cell Biol. 2020, 219. [Google Scholar] [CrossRef] [PubMed]
- Jentsch, J.A.; Kiburu, I.; Pandey, K.; Timme, M.; Ramlall, T.; Levkau, B.; Wu, J.; Eliezer, D.; Boudker, O.; Menon, A.K. Structural basis of sterol binding and transport by a yeast StARkin domain. J. Biol. Chem. 2018, 293, 5522–5531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Im, Y.J.; Raychaudhuri, S.; Prinz, W.A.; Hurley, J.H. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature 2005, 437, 154–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chowdhury, S.; Otomo, C.; Leitner, A.; Ohashi, K.; Aebersold, R.; Lander, G.C.; Otomo, T. Insights into autophagosome biogenesis from structural and biochemical analyses of the ATG2A-WIPI4 complex. Proc. Natl. Acad. Sci. USA 2018, 115, E9792–E9801. [Google Scholar] [CrossRef] [Green Version]
- Osawa, T.; Kotani, T.; Kawaoka, T.; Hirata, E.; Suzuki, K.; Nakatogawa, H.; Ohsumi, Y.; Noda, N.N. Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat. Struct. Mol. Biol. 2019, 26, 281–288. [Google Scholar] [CrossRef]
- Maeda, S.; Otomo, C.; Otomo, T. The autophagic membrane tether ATG2A transfers lipids between membranes. Elife 2019, 8, e45777. [Google Scholar] [CrossRef]
- Gómez-Sánchez, R.; Rose, J.; Guimarães, R.; Mari, M.; Papinski, D.; Rieter, E.; Geerts, W.J.; Hardenberg, R.; Kraft, C.; Ungermann, C.; et al. Atg9 establishes Atg2-dependent contact sites between the endoplasmic reticulum and phagophores. J. Cell Biol. 2018, 217, 2743–2763. [Google Scholar] [CrossRef] [PubMed]
- Velikkakath, A.K.G.; Nishimura, T.; Oita, E.; Ishihara, N.; Mizushima, N. Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol. Biol. Cell 2012, 23, 896–909. [Google Scholar] [CrossRef]
- Valverde, D.P.; Yu, S.; Boggavarapu, V.; Kumar, N.; Lees, J.A.; Walz, T.; Reinisch, K.M.; Melia, T.J. ATG2 transports lipids to promote autophagosome biogenesis. J. Cell Biol. 2019, 218, 1787–1798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawano, S.; Tamura, Y.; Kojima, R.; Bala, S.; Asai, E.; Michel, A.H.; Kornmann, B.; Riezman, I.; Riezman, H.; Sakae, Y.; et al. Structure-function insights into direct lipid transfer between membranes by Mmm1-Mdm12 of ERMES. J. Cell Biol. 2018, 217, 959–974. [Google Scholar] [CrossRef] [PubMed]
- Kotani, T.; Kirisako, H.; Koizumi, M.; Ohsumi, Y.; Nakatogawa, H. The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc. Natl. Acad. Sci. USA 2018, 115, 10363–10368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef]
- Di Mattia, T.; Martinet, A.; Ikhlef, S.; McEwen, A.G.; Nominé, Y.; Wendling, C.; Poussin-Courmontagne, P.; Voilquin, L.; Eberling, P.; Ruffenach, F.; et al. FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts. EMBO J. 2020, 39, e104369. [Google Scholar] [CrossRef] [PubMed]
- Dziurdzik, S.K.; Bean, B.D.M.; Davey, M.; Conibear, E. A VPS13D spastic ataxia mutation disrupts the conserved adaptor-binding site in yeast Vps13. Hum. Mol. Genet. 2020, 29, 635–648. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varnay, I.; Truffault, V.; Djuranovic, S.; Ursinus, A.; Coles, M.; Kessler, H. Optimized Measurement Temperature Gives Access to the Solution Structure of a 49 kDa Homohexameric-Propeller. J. Am. Chem. Soc. 2010, 132, 15692–15698. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Min, J. Structure and function of WD40 domain proteins. Protein Cell 2011, 2, 202–214. [Google Scholar] [CrossRef] [PubMed]
- Dobson-Stone, C.; Danek, A.; Rampoldi, L.; Hardie, R.; Chalmers, R.; Wood, N.; Bohlega, S.; Dotti, M.; Federico, A.; Shizuka, M.; et al. Mutational spectrum of the CHAC gene in patients with chorea-acanthocytosis. Eur. J. Hum. Genet. 2002, 10, 773–781. [Google Scholar] [CrossRef] [Green Version]
- Velayos-Baeza, A.; Levecque, C.; Dobson-Stone, C.; Monaco, A.P. The Function of Chorein. In Neuroacanthocytosis Syndromes II.; Walker, R.H., Saiki, S., Danek, A., Eds.; Springer Publishing: Berlin/Heidelberg, Germany, 2008; pp. 87–106. [Google Scholar]
- Park, J.S.; Neiman, A.M. XK is a partner for VPS13A: A molecular link between Chorea-Acanthocytosis and McLeod Syndrome. Mol. Biol. Cell 2020, 31, 2425–2436. [Google Scholar] [CrossRef] [PubMed]
- Seifert, W.; Kühnisch, J.; Maritzen, T.; Lommatzsch, S.; Hennies, H.C.; Bachmann, S.; Horn, D.; Haucke, V. Cohen syndrome-associated protein COH1 physically and functionally interacts with the small GTPase RAB6 at the Golgi complex and directs neurite outgrowth. J. Biol. Chem. 2015, 290, 3349–3358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolakowski, D.; Kaminska, J.; Zoladek, T. The binding of the APT1 domains to phosphoinositides is regulated by metal ions in vitro. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183349. [Google Scholar] [CrossRef] [PubMed]
- Kaminska, J.; Rzepnikowska, W.; Polak, A.; Flis, K.; Soczewka, P.; Bala, K.; Sienko, M.; Grynberg, M.; Kaliszewski, P.; Urbanek, A.; et al. Phosphatidylinositol-3-phosphate regulates response of cells to proteotoxic stress. Int. J. Biochem. Cell Biol. 2016, 79, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Drin, G.; Casella, J.-F.; Gautier, R.; Boehmer, T.; Schwartz, T.U.; Antonny, B. A general amphipathic α-helical motif for sensing membrane curvature. Nat. Struct. Mol. Biol. 2007, 14, 138–146. [Google Scholar] [CrossRef]
- Giménez-Andrés, M.; Čopič, A.; Antonny, B. The many faces of amphipathic helices. Biomolecules 2018, 8, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, N.; Nishimura, T.; Sakamaki, Y.; Koyama-Honda, I.; Yamamoto, H.; Mizushima, N. Differential requirement for ATG2A domains for localization to autophagic membranes and lipid droplets. FEBS Lett. 2017, 591, 3819–3830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Z.; Takahashi, Y.; He, H.; Hattori, T.; Chen, C.; Liang, X.; Chen, H.; Young, M.M.; Wang, H.G. TOM40 Targets Atg2 to Mitochondria-Associated ER Membranes for Phagophore Expansion. Cell Rep. 2019, 28, 1744–1757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Söding, J.; Biegert, A.; Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005, 33, W244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fidler, D.R.; Murphy, S.E.; Courtis, K.; Antonoudiou, P.; El-Tohamy, R.; Ient, J.; Levine, T.P. Using HHsearch to tackle proteins of unknown function: A pilot study with PH domains. Traffic 2016, 17, 1214–1226. [Google Scholar] [CrossRef] [PubMed]
- Blomberg, N.; Baraldi, E.; Nilges, M.; Saraste, M. The PH superfold: A structural scaffold for multiple functions. Trends Biochem. Sci. 1999, 24, 441–445. [Google Scholar] [CrossRef]
- Tomiyasu, A.; Nakamura, M.; Ichiba, M.; Ueno, S.; Saiki, S.; Morimoto, M.; Kobal, J.; Kageyama, Y.; Inui, T.; Wakabayashi, K.; et al. Novel pathogenic mutations and copy number variations in the VPS13A Gene in patients with chorea-acanthocytosis. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2011, 156, 620–631. [Google Scholar] [CrossRef]
- Roberts, P.; Moshitch-Moshkovitz, S.; Kvam, E.; O’Toole, E.; Winey, M.; Goldfarb, D.S. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell 2003, 14, 129–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kvam, E.; Goldfarb, D.S. Nucleus-Vacuole Junctions and Piecemeal Microautophagy of the Nucleus in S. cerevisiae. Autophagy 2007, 3, 85–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hariri, H.; Rogers, S.; Ugrankar, R.; Liu, Y.L.; Feathers, J.R.; Henne, W.M. Lipid droplet biogenesis is spatially coordinated at ER –vacuole contacts under nutritional stress. EMBO Rep. 2018, 19, 57–72. [Google Scholar] [CrossRef]
- Hariri, H.; Speer, N.; Bowerman, J.; Rogers, S.; Fu, G.; Reetz, E.; Datta, S.; Feathers, J.R.; Ugrankar, R.; Nicastro, D.; et al. Mdm1 maintains endoplasmic reticulum homeostasis by spatially regulating lipid droplet biogenesis. J. Cell Biol. 2019, 218, 1319–1334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lackner, L.L. The Expanding and Unexpected Functions of Mitochondria Contact Sites. Trends Cell Biol. 2019, 29, 580–590. [Google Scholar] [CrossRef] [PubMed]
- González Montoro, A.; Auffarth, K.; Hönscher, C.; Bohnert, M.; Becker, T.; Warscheid, B.; Reggiori, F.; van der Laan, M.; Fröhlich, F.; Ungermann, C. Vps39 Interacts with Tom40 to Establish One of Two Functionally Distinct Vacuole-Mitochondria Contact Sites. Dev. Cell 2018, 45, 621–636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.-S.; Okumura, Y.; Tachikawa, H.; Neiman, A.M. SPO71 encodes a developmental stage-specific partner for Vps13 in Saccharomyces cerevisiae. Eukaryot. Cell 2013, 12, 1530–1537. [Google Scholar] [CrossRef] [Green Version]
- Hsu, T.H.; Chen, R.H.; Cheng, Y.H.; Wang, C.W.; Riezman, H. Lipid droplets are central organelles for meiosis II progression during yeast sporulation. Mol. Biol. Cell 2017, 28, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Schütter, M.; Giavalisco, P.; Brodesser, S.; Graef, M. Local Fatty Acid Channeling into Phospholipid Synthesis Drives Phagophore Expansion during Autophagy. Cell 2020, 180, 135–149. [Google Scholar] [CrossRef] [PubMed]
- Andrejeva, G.; Gowan, S.; Lin, G.; Wong Te Fong, A.C.L.; Shamsaei, E.; Parkes, H.G.; Mui, J.; Raynaud, F.I.; Asad, Y.; Vizcay-Barrena, G.; et al. De novo phosphatidylcholine synthesis is required for autophagosome membrane formation and maintenance during autophagy. Autophagy 2019, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Dupont, N.; Chauhan, S.; Arko-Mensah, J.; Castillo, E.F.; Masedunskas, A.; Weigert, R.; Robenek, H.; Proikas-Cezanne, T.; Deretic, V. Neutral lipid stores and lipase PNPLA5 contribute to autophagosome biogenesis. Curr. Biol. 2014, 24, 609–620. [Google Scholar] [CrossRef] [Green Version]
- Shpilka, T.; Welter, E.; Borovsky, N.; Amar, N.; Mari, M.; Reggiori, F.; Elazar, Z. Lipid droplets and their component triglycerides and steryl esters regulate autophagosome biogenesis. EMBO J. 2015, 34, 2117–2131. [Google Scholar] [CrossRef] [Green Version]
- Kannan, M.; Lahiri, S.; Liu, L.K.; Choudhary, V.; Prinz, W.A. Phosphatidylserine synthesis at membrane contact sites promotes its transport out of the ER. J. Lipid Res. 2017, 58, 553–562. [Google Scholar] [CrossRef] [Green Version]
- Muñoz-Braceras, S.; Calvo, R.; Escalante, R. TipC and the chorea-acanthocytosis protein VPS13A regulate autophagy in Dictyostelium and human HeLa cells. Autophagy 2015, 11, 918–927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vonk, J.J.; Yeshaw, W.M.; Pinto, F.; Faber, A.I.E.; Lahaye, L.L.; Kanon, B.; van der Zwaag, M.; Velayos-Baeza, A.; Freire, R.; van IJzendoorn, S.C.; et al. Drosophila Vps13 Is Required for Protein Homeostasis in the Brain. PLoS One 2017, 12, e0170106. [Google Scholar] [CrossRef]
- Lupo, F.; Tibaldi, E.; Matte, A.; Sharma, A.K.; Brunati, A.M.; Alper, S.L.; Zancanaro, C.; Benati, D.; Siciliano, A.; Bertoldi, M.; et al. A new molecular link between defective autophagy and erythroid abnormalities in chorea-acanthocytosis. Blood 2016, 128, 2976–2987. [Google Scholar] [CrossRef] [Green Version]
- Stewart, G.W.; Wilmore, S.M.S.; Ohno, S.; Terada, N. Questions of Cell Shape. In Neuroacanthocytosis Syndromes II.; Walker, R.H., Saiki, S., Danek, A., Eds.; Springer-Verlag Berlin: Heidelberg, Germany, 2008; pp. 115–132. ISBN 9783540716938. [Google Scholar]
- Redman, C.; Huima, T.; Robbins, E.; Lee, S.; Marsh, W. Effect of phosphatidylserine on the shape of McLeod red cell acanthocytes. Blood 1989, 74, 1826–1835. [Google Scholar] [CrossRef] [Green Version]
- Yang, R.-Y.; Xue, H.; Yu, L.; Velayos-Baeza, A.; Monaco, A.P.; Liu, F.-T. Identification of VPS13C as a galectin-12-binding protein that regulates galectin-12 protein stability and adipogenesis. PLoS One 2016, 11. [Google Scholar] [CrossRef] [Green Version]
- Ramseyer, V.D.; Kimler, V.A.; Granneman, J.G. Vacuolar protein sorting 13C is a novel lipid droplet protein that inhibits lipolysis in brown adipocytes. Mol. Metab. 2018, 7, 57–70. [Google Scholar] [CrossRef]
- Elbaz-Alon, Y.; Eisenberg-Bord, M.; Shinder, V.; Stiller, S.B.; Shimoni, E.; Wiedemann, N.; Geiger, T.; Schuldiner, M. Lam6 Regulates the Extent of Contacts between Organelles. Cell Rep. 2015, 12, 7–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matoba, K.; Kotani, T.; Tsutsumi, A.; Tsuji, T.; Mori, T.; Noshiro, D.; Sugita, Y.; Nomura, N.; Iwata, S.; Ohsumi, Y.; et al. Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat. Struct. Mol. Biol. 2020, 1–9. [Google Scholar] [CrossRef]
- Suzuki, J.; Denning, D.P.; Imanishi, E.; Horvitz, H.R.; Nagata, S. Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science 2013, 341, 403–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, J.; Imanishi, E.; Nagata, S. Exposure of phosphatidylserine by Xkrelated protein family members during apoptosis. J. Biol. Chem. 2014, 289, 30257–30267. [Google Scholar] [CrossRef] [Green Version]
- Roulis, E.; Hyland, C.; Flower, R.; Gassner, C.; Jung, H.H.; Frey, B.M. Molecular Basis and Clinical Overview of McLeod Syndrome Compared with Other Neuroacanthocytosis Syndromes: A Review. JAMA Neurol. 2018, 75, 1554–1562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huttlin, E.L.; Ting, L.; Bruckner, R.J.; Gebreab, F.; Gygi, M.P.; Szpyt, J.; Tam, S.; Zarraga, G.; Colby, G.; Baltier, K.; et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 2015, 162, 425–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Homolog | Localization | Reference |
|---|---|---|
| VPS13A | Endoplasmic reticulum-mitochondria | [15,16,20] |
| Endoplasmic reticulum-lipid droplet | [15,16] | |
| Mitochondria-endosome | [20] | |
| VPS13B | Golgi apparatus | [21] |
| Endosome | [17] | |
| Acrosome | [22] | |
| VPS13C | Endoplasmic reticulum-endosome/lysosome | [15] |
| Endoplasmic reticulum-lipid droplet | [15] | |
| Phagophore | [23,24,25] | |
| VPS13D | Mitochondria | [26] |
| Golgi apparatus | [26] | |
| Peroxisome | [26] |
| Membrane Contact Site | Reference |
|---|---|
| Nucleus-vacuole junctions (NVJs) | [18,27,28] |
| Mitochondria-endosome | [28] |
| Vacuole-mitochondria patch (vCLAMP) | [27,28] |
| Single Organelle | Reference |
| Endosome | [18,29,30] |
| Mitochondria | [27,28,31] |
| Vacuole | [18,27,28,32] |
| Prospore membrane | [29] |
| Golgi apparatus1 | [33] |
| Peroxisome | [31] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dziurdzik, S.K.; Conibear, E. The Vps13 Family of Lipid Transporters and Its Role at Membrane Contact Sites. Int. J. Mol. Sci. 2021, 22, 2905. https://doi.org/10.3390/ijms22062905
Dziurdzik SK, Conibear E. The Vps13 Family of Lipid Transporters and Its Role at Membrane Contact Sites. International Journal of Molecular Sciences. 2021; 22(6):2905. https://doi.org/10.3390/ijms22062905
Chicago/Turabian StyleDziurdzik, Samantha Katarzyna, and Elizabeth Conibear. 2021. "The Vps13 Family of Lipid Transporters and Its Role at Membrane Contact Sites" International Journal of Molecular Sciences 22, no. 6: 2905. https://doi.org/10.3390/ijms22062905
APA StyleDziurdzik, S. K., & Conibear, E. (2021). The Vps13 Family of Lipid Transporters and Its Role at Membrane Contact Sites. International Journal of Molecular Sciences, 22(6), 2905. https://doi.org/10.3390/ijms22062905