Engineering a Novel Bivalent Oral Vaccine against Enteric Fever
Abstract
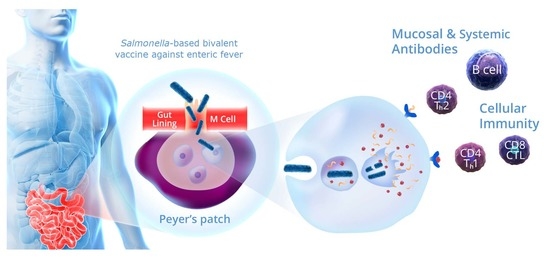
:1. Introduction
2. Results
2.1. Converting Flagellin from H:d to H:a
2.2. Modifying LPS from O:9 to O:2
2.3. Constructing the Final New Strain, ZH9PA
2.4. Evaluating the Growth of ZH9PA
2.5. Evaluating the Immunogenicity of the Basic Bivalent Enteric Fever Vaccine
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Media
4.2. Generation of Dif-Flanked Antibiotic Resistance Gene Insertion Cassette
4.3. Chromosomal Integration Procedure
4.4. Conversion of Flagellin
4.5. Immunostaining
4.6. Western and Dot Blot
4.7. Silver Staining
4.8. Growth Studies
4.9. Murine Immunogenicity
4.10. ELISA
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parry, C.M.; Ribeiro, I.; Walia, K.; Rupali, P.; Baker, S.; Basnyat, B. Multidrug resistant enteric fever in South Asia: Unmet medical needs and opportunities. BMJ 2019, 364, k5322. [Google Scholar] [CrossRef] [Green Version]
- GBD. The global burden of typhoid and paratyphoid fevers: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect. Dis. 2019, 19, 369–381. [Google Scholar] [CrossRef] [Green Version]
- Kariuki, S.; Gordon, M.A.; Feasey, N.; Parry, C.M. Antimicrobial resistance and management of invasive Salmonella disease. Vaccine 2015, 33 (Suppl. 3), C21–C29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akram, J.; Khan, A.S.; Khan, H.A.; Gilani, S.A.; Akram, S.J.; Ahmad, F.J.; Mehboob, R. Extensively Drug-Resistant (XDR) Typhoid: Evolution, Prevention, and Its Management. Biomed Res. Int. 2020, 2020, 6432580. [Google Scholar] [CrossRef] [PubMed]
- WHO. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery and Development of New Antibiotics; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Anwar, E.; Goldberg, E.; Fraser, A.; Acosta, C.J.; Paul, M.; Leibovici, L. Vaccines for preventing typhoid fever. Cochrane Database Syst. Rev. 2014. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.B. Vaccines for typhoid fever and other salmonelloses. Curr. Opin. Infect. Dis. 2012, 25, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Sahastrabuddhe, S.; Carbis, R.; Wierzba, T.F.; Ochiai, R.L. Increasing rates of Salmonella Paratyphi A and the current status of its vaccine development. Expert Rev. Vaccines 2013, 12, 1021–1031. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, Y.K.; Adedare, T.A.; Ehinmidu, J.O. Antibiotic sensitivity profiles of salmonella organisms isolated from presumptive typhoid patients in Zaria, northern Nigeria. Afr. J. Med. Med. Sci. 2005, 34, 109–114. [Google Scholar] [PubMed]
- Akinyemi, K.O.; Bamiro, B.S.; Coker, A.O. Salmonellosis in Lagos, Nigeria: Incidence of Plasmodium falciparum-associated co-infection, patterns of antimicrobial resistance, and emergence of reduced susceptibility to fluoroquinolones. J. Health Popul. Nutr. 2007, 25, 351–358. [Google Scholar] [PubMed]
- Antillón, M.; Warren, J.L.; Crawford, F.W.; Weinberger, D.M.; Kürüm, E.; Pak, G.D.; Marks, F.; Pitzer, V.E. The burden of typhoid fever in low- and middle-income countries: A meta-regression approach. PLoS Negl. Trop. Dis. 2017, 11, e0005376. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, K.S.; Pak, G.D.; Excler, J.L.; Sahastrabuddhe, S.; Marks, F.; Kim, J.H.; Mogasale, V. Spatial and Temporal Patterns of Typhoid and Paratyphoid Fever Outbreaks: A Worldwide Review, 1990–2018. Clin. Infect. Dis. 2019, 69, S499–S509. [Google Scholar] [CrossRef]
- Guzman, C.A.; Borsutzky, S.; Griot-Wenk, M.; Metcalfe, I.C.; Pearman, J.; Collioud, A.; Favre, D.; Dietrich, G. Vaccines against typhoid fever. Vaccine 2006, 24, 3804–3811. [Google Scholar] [CrossRef]
- Roland, K.L.; Tinge, S.A.; Kochi, S.K.; Thomas, L.J.; Killeen, K.P. Reactogenicity and immunogenicity of live attenuated Salmonella enterica serovar Paratyphi A enteric fever vaccine candidates. Vaccine 2010, 28, 3679–3687. [Google Scholar] [CrossRef] [PubMed]
- Wahid, R.; Kotloff, K.L.; Levine, M.M.; Sztein, M.B. Cell mediated immune responses elicited in volunteers following immunization with candidate live oral Salmonella enterica serovar Paratyphi A attenuated vaccine strain CVD 1902. Clin. Immunol. 2019, 201, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Darton, T.C.; Jones, C.; Blohmke, C.J.; Waddington, C.S.; Zhou, L.; Peters, A.; Haworth, K.; Sie, R.; Green, C.A.; Jeppesen, C.A.; et al. Using a Human Challenge Model of Infection to Measure Vaccine Efficacy: A Randomised, Controlled Trial Comparing the Typhoid Vaccines M01ZH09 with Placebo and Ty21a. PLoS Negl. Trop. Dis. 2016, 10, e0004926. [Google Scholar] [CrossRef] [PubMed]
- Hindle, Z.; Chatfield, S.N.; Phillimore, J.; Bentley, M.; Johnson, J.; Cosgrove, C.A.; Ghaem-Maghami, M.; Sexton, A.; Khan, M.; Brennan, F.R.; et al. Characterization of Salmonella enterica derivatives harboring defined aroC and Salmonella pathogenicity island 2 type III secretion system (ssaV) mutations by immunization of healthy volunteers. Infect. Immun. 2002, 70, 3457–3467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirkpatrick, B.D.; McKenzie, R.; O’Neill, J.P.; Larsson, C.J.; Bourgeois, A.L.; Shimko, J.; Bentley, M.; Makin, J.; Chatfield, S.; Hindle, Z.; et al. Evaluation of Salmonella enterica serovar Typhi (Ty2 aroC-ssaV-) M01ZH09, with a defined mutation in the Salmonella pathogenicity island 2, as a live, oral typhoid vaccine in human volunteers. Vaccine 2006, 24, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, B.D.; Tenney, K.M.; Larsson, C.J.; O’Neill, J.P.; Ventrone, C.; Bentley, M.; Upton, A.; Hindle, Z.; Fidler, C.; Kutzko, D.; et al. The novel oral typhoid vaccine M01ZH09 is well tolerated and highly immunogenic in 2 vaccine presentations. J. Infect. Dis. 2005, 192, 360–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyon, C.E.; Sadigh, K.S.; Carmolli, M.P.; Harro, C.; Sheldon, E.; Lindow, J.C.; Larsson, C.J.; Martinez, T.; Feller, A.; Ventrone, C.H.; et al. In a randomized, double-blinded, placebo-controlled trial, the single oral dose typhoid vaccine, M01ZH09, is safe and immunogenic at doses up to 1.7 × 10(10) colony-forming units. Vaccine 2010, 28, 3602–3608. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.H.; Nguyen, T.D.; Nguyen, T.T.; Ninh, T.T.; Tran, N.B.; Nguyen, V.M.; Tran, T.T.; Cao, T.T.; Pham, V.M.; Nguyen, T.C.; et al. A randomised trial evaluating the safety and immunogenicity of the novel single oral dose typhoid vaccine M01ZH09 in healthy Vietnamese children. PLoS ONE 2010, 5, e11778. [Google Scholar] [CrossRef]
- Prokarium. Study to Evaluate the Safety, Tolerability and Immunogenicity of a Potential Enteric Fever Vaccine. 2019. Available online: https://ClinicalTrials.gov/show/NCT04349553 (accessed on 27 February 2021).
- Bloor, A.E.; Cranenburgh, R.M. An efficient method of selectable marker gene excision by Xer recombination for gene replacement in bacterial chromosomes. Appl. Environ. Microbiol. 2006, 72, 2520–2525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grimont, P.A.D.; Weill, F.X. Antigenic Formulae of the Salmonella Serovars; WHO Collaborating Centre for Reference and Research on Salmonella, Institut Pasteur: Paris, France, 2007. [Google Scholar]
- Liu, Q.; Li, P.; Luo, H.; Curtiss, R., 3rd; Kong, Q. Attenuated Salmonella Typhimurium expressing Salmonella Paratyphoid A O-antigen induces protective immune responses against two Salmonella strains. Virulence 2019, 10, 82–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.; Cole, R.A.; Reeves, P.R. An O-antigen processing function for Wzx (RfbX): A promising candidate for O-unit flippase. J. Bacteriol. 1996, 178, 2102–2107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fitzgerald, C.; Sherwood, R.; Gheesling, L.L.; Brenner, F.W.; Fields, P.I. Molecular analysis of the rfb O antigen gene cluster of Salmonella enterica serogroup O:6,14 and development of a serogroup-specific PCR assay. Appl. Environ. Microbiol. 2003, 69, 6099–6105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, Y.; Cunneen, M.M.; Reeves, P.R. The Wzx translocases for Salmonella enterica O-antigen processing have unexpected serotype specificity. Mol. Microbiol. 2012, 84, 620–630. [Google Scholar] [CrossRef] [PubMed]
- MacLennan, C.A.; Martin, L.B.; Micoli, F. Vaccines against invasive Salmonella disease: Current status and future directions. Hum. Vaccines Immunother. 2014, 10, 1478–1493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hajam, I.A.; Dar, P.A.; Shahnawaz, I.; Jaume, J.C.; Lee, J.H. Bacterial flagellin-a potent immunomodulatory agent. Exp. Mol. Med. 2017, 49, e373. [Google Scholar] [CrossRef] [PubMed]
- Nagy, G.; Pál, T. Lipopolysaccharide: A tool and target in enterobacterial vaccine development. Biol. Chem. 2008, 389, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Park, B.S.; Lee, J.O. Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp. Mol. Med. 2013, 45, e66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colwell, D.E.; Michalek, S.M.; Briles, D.E.; Jirillo, E.; McGhee, J.R. Monoclonal antibodies to Salmonella lipopolysaccharide: Anti-O-polysaccharide antibodies protect C3H mice against challenge with virulent Salmonella typhimurium. J. Immunol. 1984, 133, 950–957. [Google Scholar]
- Forbes, S.J.; Martinelli, D.; Hsieh, C.; Ault, J.G.; Marko, M.; Mannella, C.A.; Mantis, N.J. Association of a protective monoclonal IgA with the O antigen of Salmonella enterica serovar Typhimurium impacts type 3 secretion and outer membrane integrity. Infect. Immun. 2012, 80, 2454–2463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michetti, P.; Mahan, M.J.; Slauch, J.M.; Mekalanos, J.J.; Neutra, M.R. Monoclonal secretory immunoglobulin A protects mice against oral challenge with the invasive pathogen Salmonella typhimurium. Infect. Immun. 1992, 60, 1786–1792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eom, J.S.; Seok Kim, J.; Im Jang, J.; Kim, B.H.; Young Yoo, S.; Hyeon Choi, J.; Bang, I.S.; Lee, I.S.; Keun Park, Y. Enhancement of host immune responses by oral vaccination to Salmonella enterica serovar Typhimurium harboring both FliC and FljB flagella. PLoS ONE 2013, 8, e74850. [Google Scholar] [CrossRef] [PubMed]
- Simon, R.; Tennant, S.M.; Wang, J.Y.; Schmidlein, P.J.; Lees, A.; Ernst, R.K.; Pasetti, M.F.; Galen, J.E.; Levine, M.M. Salmonella enterica serovar enteritidis core O polysaccharide conjugated to H:g,m flagellin as a candidate vaccine for protection against invasive infection with S. enteritidis. Infect. Immun. 2011, 79, 4240–4249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bobat, S.; Flores-Langarica, A.; Hitchcock, J.; Marshall, J.L.; Kingsley, R.A.; Goodall, M.; Gil-Cruz, C.; Serre, K.; Leyton, D.L.; Letran, S.E.; et al. Soluble flagellin, FliC, induces an Ag-specific Th2 response, yet promotes T-bet-regulated Th1 clearance of Salmonella typhimurium infection. Eur. J. Immunol. 2011, 41, 1606–1618. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Knirel, Y.A.; Feng, L.; Perepelov, A.V.; Senchenkova, S.N.; Reeves, P.R.; Wang, L. Structural diversity in Salmonella O antigens and its genetic basis. FEMS Microbiol. Rev. 2014, 38, 56–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- HSE. The Approved List of Biological Agents: Advisory Committee on Dangerous Pathogens; Health and Safety Executive: Bootle, UK, 2013. [Google Scholar]
- Boudehen, Y.M.; Wallat, M.; Rousseau, P.; Neyrolles, O.; Gutierrez, C. An improved Xer-cise technology for the generation of multiple unmarked mutants in Mycobacteria. Biotechniques 2020, 68, 106–110. [Google Scholar] [CrossRef] [PubMed]
- GraphPad-Prism. Software for Data Analysis and Graphing Solutions, Version 9; GraphPad Software: San Diego, CA, USA, 2020. [Google Scholar]





| Strain or Plasmid | Description | Source or Reference |
|---|---|---|
| Strains | ||
| Top10 E. coli | F– mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK λ– rpsL(StrR) endA1 nupG | Life Technologies, Paisley, UK |
| S. Typhi ZH9 | Ty2 ΔaroC ΔssaV | Prokarium |
| S. Typhi ZH9PF | Ty2 ΔaroC ΔssaV Δt0918::SPA0911 | This work |
| S. Typhi ZH9PL2 | Ty2 ΔaroC ΔssaV ΔrfbE | This work |
| S. Typhi ZH9PA | Ty2 ΔaroC ΔssaV ΔrfbE::wbdR Δt0918::SPA0911 | This work |
| Plasmids | ||
| pBRT1Nc | Synthesised with Chloramphenical resistance gene | This work |
| pUCFlic2 | Synthesised S. Paratyphi A fliC | This work |
| pUCWbdR | Synthesised E. coli wbdR | This work |
| pUCpF-difcat | Precursor S. Paratyphi A fliC integration Xer-cise plasmid | This work |
| pUCpW-difcat | Precursor wbdR integration Xer-cise plasmid | This work |
| pL2-difcat | Precursor rfbE deletion Xer-cise cassette | This work |
| pLGBK | Lambda Red helper plasmid | Prokarium |
| Primer Name | Primer Sequence (5′-3′) | Primer Function |
|---|---|---|
| 5NotIdifcat | taagcggccgcATTTAACATAATATACATTATGCGCACCgcccgaacaccac | Primers designed to include NotI restriction sites at the 5′ and 3′ end respectively. The lowercase letters represent the region of homology to pBRT1Nc; the NotI restriction site is underlined and dif sites are in capital letters |
| 3NotIdifcat | ggcggccgcGGTGCGCATAATGTATATTATGTTAAATgggcgagtttacatctcaaaaccg | |
| rfbE del F | AATAGGATGGAAAAGAGAGTTCTCTCTTGTTGATGCATTAACTGAAATAATTGAAGAGGAAGGGAAATGAAAAGCTTGGTACCGAGCTCG | rfbE deletion |
| rfbE del R | TTTGAAAGCCAAGAGGAAGCGGCAATAATAAGATGTCTTGGAATTCTAACCAACCTCAGTTTCCTCACTCTAGATGCATGCTCGAGCGGC | rfbE deletion |
| L1 | AGGCTTGACTACAGAGCATTTAGATTATGTAG | Diagnostic primer for LPS locus |
| L6 | ACATACTTCTACAATTAAGGAGTGAGAAGATTGATTATTAATACT | Diagnostic primer for LPS locus |
| L2 | TCACGACTTACATCCTACTTCG | Diagnostic primer for LPS locus |
| L3 | TGTTCCTGCCGGTATAACTG | Diagnostic primer for LPS locus |
| L4 | CAGTTTCCTCACGTCAGCTT | Diagnostic primer for LPS locus |
| L5 | CTGGCCATAATGCTTGTAATACCGCA | Diagnostic primer for LPS locus |
| F1 | GCTGACTTGCGCATAAGCTTTGA | Diagnostic primer for Flagellin locus |
| F8 | AACAGCCCTGCGTTAAATGAGT | Diagnostic primer for Flagellin locus |
| F3 | TATTGCTCTGACGCTCAATG | Diagnostic primer for Flagellin locus |
| F6 | ACGGTGATTTCTTTCATTACACAG | Diagnostic primer for Flagellin locus |
| F5 | TTCAGCAGTATCAGCGCCGGT | Diagnostic primer for Flagellin locus |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soulier, A.; Prevosto, C.; Chol, M.; Deban, L.; Cranenburgh, R.M. Engineering a Novel Bivalent Oral Vaccine against Enteric Fever. Int. J. Mol. Sci. 2021, 22, 3287. https://doi.org/10.3390/ijms22063287
Soulier A, Prevosto C, Chol M, Deban L, Cranenburgh RM. Engineering a Novel Bivalent Oral Vaccine against Enteric Fever. International Journal of Molecular Sciences. 2021; 22(6):3287. https://doi.org/10.3390/ijms22063287
Chicago/Turabian StyleSoulier, Annelise, Claudia Prevosto, Mary Chol, Livija Deban, and Rocky M. Cranenburgh. 2021. "Engineering a Novel Bivalent Oral Vaccine against Enteric Fever" International Journal of Molecular Sciences 22, no. 6: 3287. https://doi.org/10.3390/ijms22063287
APA StyleSoulier, A., Prevosto, C., Chol, M., Deban, L., & Cranenburgh, R. M. (2021). Engineering a Novel Bivalent Oral Vaccine against Enteric Fever. International Journal of Molecular Sciences, 22(6), 3287. https://doi.org/10.3390/ijms22063287