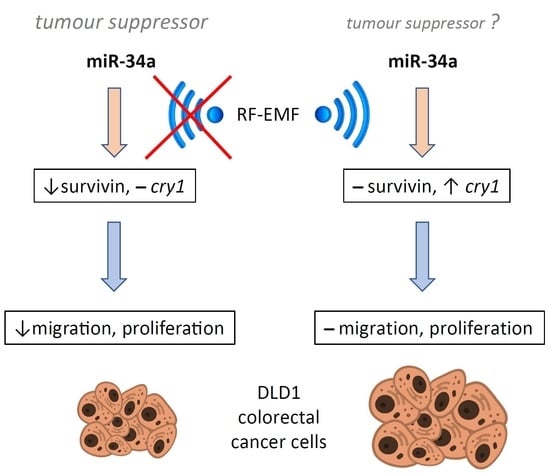
2.4 GHz Electromagnetic Field Influences the Response of the Circadian Oscillator in the Colorectal Cancer Cell Line DLD1 to miR-34a-Mediated Regulation
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aschoff, J. Biological rhythms. In Handbook of Behavioral Neurobiology; Dethier, V.G., Goy, R.W., Eds.; Platinum Press: New York, NY, USA; London, UK, 1981; Volume 4, p. 545. [Google Scholar] [CrossRef]
- Pilorz, V.; Astiz, M.; Heinen, K.O.; Rawashdeh, O.; Oster, H. The Concept of Coupling in the Mammalian Circadian Clock Network. J. Mol. Biol. 2020, 432, 3618–3638. [Google Scholar] [CrossRef] [PubMed]
- Patke, A.; Young, M.W.; Axelrod, S. Molecular mechanisms and physiological importance of circadian rhythms. Nat. Rev. Mol. Cell Biol. 2020, 21, 67–84. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Lazar, M.A. Transcriptional control of circadian rhythms and metabolism: A matter of time and space. Endocr. Rev. 2020, 41, 707–732. [Google Scholar] [CrossRef] [PubMed]
- Pegoraro, M.; Tauber, E. The role of microRNAs (miRNA) in circadian rhythmicity. J. Genet. 2008, 87, 505–511. [Google Scholar] [CrossRef] [Green Version]
- Hansen, K.F.; Sakamoto, K.; Obrietan, K. MicroRNAs: A potential interface between the circadian clock and human health. Genome Med. 2011, 17, 10. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tian, G.; Li, Z.; Zheng, L. The crosstalk between miRNA and mammalian circadian clock. Curr. Med. Chem. 2015, 22, 1582–1588. [Google Scholar] [CrossRef] [PubMed]
- Anna, G.; Kannan, N.N. Post-transcriptional modulators and mediators of the circadian clock. Chronobiol. Int. 2021, 38, 1244–1261. [Google Scholar] [CrossRef]
- Ray, I.; Goswami, S. Circadian rhythm genes in cancer: Insight into their functions and regulation involving noncoding RNAs. Chronobiol. Int. 2021, 38, 1231–1243. [Google Scholar] [CrossRef]
- Cheng, H.Y.; Papp, J.W.; Varlamova, O.; Dziema, H.; Russell, B.; Curfman, J.P.; Nakazawa, T.; Shimizu, K.; Okamura, H.; Impey, S.; et al. microRNA modulation of circadian-clock period and entrainment. Neuron 2007, 54, 813–829. [Google Scholar] [CrossRef]
- Han, Y.; Meng, F.; Venter, J.; Wu, N.; Wan, Y.; Standeford, H.; Francis, H.; Meininger, C.; Greene, J., Jr.; Trzeciakowski, J.P.; et al. miR-34a-dependent overexpression of Per1 decreases cholangiocarcinoma growth. J. Hepatol. 2016, 64, 1295–1304. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; D’Alessandro, M.; Lee, C. miRNAs are required for generating a time delay critical for the circadian oscillator. Curr. Biol. 2013, 23, 1959–1968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, X.; Sun, B.; Huang, J.; Xu, L.; Pan, J.; Fang, C.; Tao, Y.; Hu, S.; Li, R.; Han, X.; et al. The role of miR-182 in regulating pineal CLOCK expression after hypoxia-ischemia brain injury in neonatal rats. Neurosci. Lett. 2015, 591, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Zhou, L.; Yang, S.Y.; Cao, J.M. A novel role of microRNA 17-5p in the modulation of circadian rhythm. Sci. Rep. 2016, 6, 30070. [Google Scholar] [CrossRef]
- Li, A.; Lin, X.; Tan, X.; Yin, B.; Han, W.; Zhao, J.; Yuan, J.; Qiang, B.; Peng, X. Circadian gene Clock contributes to cell proliferation and migration of glioma and is directly regulated by tumor-suppressive miR-124. FEBS Lett. 2013, 587, 2455–2460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Q.; Fan, X.; Liu, Y.; Xu, L.; Dong, P.; Song, L.; Qian, R. miR-455-5p regulates circadian rhythms by accelerating the degradation of Clock mRNA. IUBMB Life 2022, 74, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, P.; Chen, S.; Zhang, Z.; Liang, T.; Liu, C. Rhythmic expression of miR-27b-3p targets the clock gene Bmal1 at the posttranscriptional level in the mouse liver. FASEB J. 2016, 30, 2151–2160. [Google Scholar] [CrossRef]
- Tan, X.; Zhang, P.; Zhou, L.; Yin, B.; Pan, H.; Peng, X. Clock-controlled mir-142-3p can target its activator, Bmal1. BMC Mol. Biol. 2012, 13, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shende, V.R.; Neuendorff, N.; Earnest, D.J. Role of miR-142-3p in the post-transcriptional regulation of the clock gene Bmal1 in the mouse SCN. PLoS ONE 2013, 8, e65300. [Google Scholar] [CrossRef]
- Curtis, A.M.; Fagundes, C.T.; Yang, G.; Palsson-McDermott, E.M.; Wochal, P.; McGettrick, A.F.; Foley, N.H.; Early, J.O.; Chen, L.; Zhang, H.; et al. Circadian control of innate immunity in macrophages by miR-155 targeting Bmal1. Proc. Natl. Acad. Sci. USA 2015, 112, 7231–7236. [Google Scholar] [CrossRef]
- Jiang, W.; Zhao, S.; Shen, J.; Guo, L.; Sun, Y.; Zhu, Y.; Ma, Z.; Zhang, X.; Hu, Y.; Xiao, W.; et al. The MiR-135b-BMAL1-YY1 loop disturbs pancreatic clockwork to promote tumourigenesis and chemoresistance. Cell Death Dis. 2018, 9, 149. [Google Scholar] [CrossRef] [Green Version]
- Bu, Y.; Yoshida, A.; Chitnis, N.; Altman, B.J.; Tameire, F.; Oran, A.; Gennaro, V.; Armeson, K.E.; McMahon, S.B.; Wertheim, G.B.; et al. A PERK-miR-211 axis suppresses circadian regulators and protein synthesis to promote cancer cell survival. Nat. Cell Biol. 2018, 20, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.H.; Kojima, S.; Shimomura, K.; Koike, N.; Buhr, E.D.; Furukawa, T.; Ko, C.H.; Gloston, G.; Ayoub, C.; Nohara, K.; et al. Period2 3′-UTR and microRNA-24 regulate circadian rhythms by repressing PERIOD2 protein accumulation. Proc. Natl. Acad. Sci. USA 2017, 114, E8855–E8864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, I.; Kim, D.; Kim, J.; Jang, S.; Choi, M.; Choe, H.K.; Choe, Y.; Kim, K. microRNA-25 as a novel modulator of circadian Period2 gene oscillation. Exp. Mol. Med. 2020, 52, 1614–1626. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Miller, C.; Miraglia, L.J.; Romero, A.; Mure, L.S.; Panda, S.; Kay, S.A. A genome-wide microRNA screen identifies the microRNA-183/96/182 cluster as a modulator of circadian rhythms. Proc. Natl. Acad. Sci. USA 2021, 118, e2020454118. [Google Scholar] [CrossRef] [PubMed]
- Daimiel-Ruiz, L.; Klett-Mingo, M.; Konstantinidou, V.; Micó, V.; Aranda, J.F.; García, B.; Martínez-Botas, J.; Dávalos, A.; Fernández-Hernando, C.; Ordovás, J.M. Dietary lipids modulate the expression of miR-107, an miRNA that regulates the circadian system. Mol. Nutr. Food Res. 2015, 59, 552–565. [Google Scholar] [CrossRef] [Green Version]
- Hasakova, K.; Vician, M.; Reis, R.; Zeman, M.; Herichova, I. Sex-dependent correlation between survival and expression of genes related to the circadian oscillator in patients with colorectal cancer. Chronobiol. Int. 2018, 35, 1423–1434. [Google Scholar] [CrossRef]
- Nagel, R.; Clijsters, L.; Agami, R. The miRNA-192/194 cluster regulates the Period gene family and the circadian clock. FEBS J. 2009, 276, 5447–5455. [Google Scholar] [CrossRef]
- Lee, K.H.; Kim, S.H.; Lee, H.R.; Kim, W.; Kim, D.Y.; Shin, J.C.; Yoo, S.H.; Kim, K.T. MicroRNA-185 oscillation controls circadian amplitude of mouse Cryptochrome 1 via translational regulation. Mol. Biol. Cell 2013, 24, 2248–2255. [Google Scholar] [CrossRef]
- Guo, X.; Zhu, Y.; Hong, X.; Zhang, M.; Qiu, X.; Wang, Z.; Qi, Z.; Hong, X. miR-181d and c-myc-mediated inhibition of CRY2 and FBXL3 reprograms metabolism in colorectal cancer. Cell Death Dis. 2017, 8, e2958. [Google Scholar] [CrossRef]
- Lewczuk, B.; Redlarski, G.; Zak, A.; Ziółkowska, N.; Przybylska-Gornowicz, B.; Krawczuk, M. Influence of electric, magnetic, and electromagnetic fields on the circadian system: Current stage of knowledge. Biomed. Res. Int. 2014, 2014, 169459. [Google Scholar] [CrossRef] [Green Version]
- Hollenbach, D.F.; Herndon, J.M. Deep-Earth reactor: Nuclear fission, helium, and the geomagnetic field. Proc. Natl. Acad. Sci. USA 2001, 98, 11085–11090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardell, L. World Health Organization, radiofrequency radiation and health—A hard nut to crack (Review). Int. J. Oncol. 2017, 51, 405–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.H.; Lee, J.K.; Kim, H.G.; Kim, K.B.; Kim, H.R. Possible Effects of Radiofrequency Electromagnetic Field Exposure on Central Nerve System. Biomol. Ther. (Seoul) 2019, 27, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Boussad, Y.; Chen, X.L.; Legout, A.; Chaintreau, A.; Dabbous, W. Longitudinal study of exposure to radio frequencies at population scale. Environ. Int. 2022, 162, 107144. [Google Scholar] [CrossRef] [PubMed]
- Mumtaz, S.; Rana, J.N.; Choi, E.H.; Han, I. Microwave Radiation and the Brain: Mechanisms, Current Status, and Future Prospects. Int. J. Mol. Sci. 2022, 23, 9288. [Google Scholar] [CrossRef]
- Qin, F.; Shen, T.; Cao, H.; Qian, J.; Zou, D.; Ye, M.; Pei, H. CeO2NPs relieve radiofrequency radiation, improve testosterone synthesis, and clock gene expression in Leydig cells by enhancing antioxidation. Int. J. Nanomed. 2019, 14, 4601–4611. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhang, H.; Zhang, Z.; Sun, B.; Tang, C.; Zhang, L.; Jiang, Z.; Ding, B.; Liao, Y.; Cai, P. Simulated mobile communication frequencies (3.5 GHz) emitted by a signal generator affects the sleep of Drosophila melanogaster. Environ. Pollut. 2021, 283, 117087. [Google Scholar] [CrossRef]
- Feillet, C.; van der Horst, G.T.; Levi, F.; Rand, D.A.; Delaunay, F. Coupling between the Circadian Clock and Cell Cycle Oscillators: Implication for Healthy Cells and Malignant Growth. Front. Neurol. 2015, 6, 96. [Google Scholar] [CrossRef] [Green Version]
- Yao, J.; He, C.; Zhao, W.; Hu, N.; Long, D. Circadian clock and cell cycle: Cancer and chronotherapy. Acta Histochem. 2021, 123, 151816. [Google Scholar] [CrossRef]
- Li, H.X. The role of circadian clock genes in tumors. Onco. Targets Ther. 2019, 12, 3645–3660. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y. Roles of circadian clocks in cancer pathogenesis and treatment. Exp. Mol. Med. 2021, 53, 1529–1538. [Google Scholar] [CrossRef] [PubMed]
- Xiang, R.; Cui, Y.; Wang, Y.; Xie, T.; Yang, X.; Wang, Z.; Li, J.; Li, Q. Circadian clock gene Per2 downregulation in non-small cell lung cancer is associated with tumour progression and metastasis. Oncol. Rep. 2018, 40, 3040–3048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hua, H.; Wang, Y.; Wan, C.; Liu, Y.; Zhu, B.; Yang, C.; Wang, X.; Wang, Z.; Cornelissen-Guillaume, G.; Halberg, F. Circadian gene mPer2 overexpression induces cancer cell apoptosis. Cancer Sci. 2006, 97, 589–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, C.M.; Huang, S.F.; Zeng, J.M.; Liu, D.B.; Xiao, Q.; Tian, W.J.; Zhu, X.D.; Huang, Z.G.; Feng, W.L. Per2 inhibits k562 leukemia cell growth in vitro and in vivo through cell cycle arrest and apoptosis induction. Pathol. Oncol. Res. 2010, 16, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, S.; Li, X.; Li, B.; Li, Y.; Xia, K.; Yang, Y.; Aman, S.; Wang, M.; Wu, H. Circadian protein BMAL1 promotes breast cancer cell invasion and metastasis by up-regulating matrix metalloproteinase9 expression. Cancer Cell Int. 2019, 16, 182. [Google Scholar] [CrossRef]
- Xiao, L.; Chang, A.K.; Zang, M.X.; Bi, H.; Li, S.; Wang, M.; Xing, X.; Wu, H. Induction of the CLOCK gene by E2-ERα signaling promotes the proliferation of breast cancer cells. PLoS ONE 2014, 9, e95878. [Google Scholar] [CrossRef]
- Mou, J.; Dai, J.Q.; Liu, M.L.; Ni, Q.R.; Zhang, Y.J.; Wen, J.; Song, Y.P. [Knockout of BMAL1 Gene Induces Apoptosis of HL-60 Cells and Inhibits its Proliferation]. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2018, 26, 1027–1032. [Google Scholar] [CrossRef]
- Tang, Q.; Cheng, B.; Xie, M.; Chen, Y.; Zhao, J.; Zhou, X.; Chen, L. Circadian Clock Gene Bmal1 Inhibits Tumorigenesis and Increases Paclitaxel Sensitivity in Tongue Squamous Cell Carcinoma. Cancer Res. 2017, 77, 532–544. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Meng, X.; Li, Y.; Liu, L.; He, Q.; Jiang, J.; Chen, Y.; Li, X.; Li, Y.; Tang, Y.; et al. Circadian clock gene BMAL1 inhibits the proliferation and tumor-formation ability of nasopharyngeal carcinoma cells and increases the sensitivity of radiotherapy. Chronobiol. Int. 2022, 39, 1340–1351. [Google Scholar] [CrossRef]
- Gwon, D.H.; Lee, W.Y.; Shin, N.; Kim, S.I.; Jeong, K.; Lee, W.H.; Kim, D.W.; Hong, J.; Lee, S.Y. BMAL1 Suppresses Proliferation, Migration, and Invasion of U87MG Cells by Down regulating Cyclin B1, Phospho-AKT, and Metalloproteinase-9. Int. J. Mol. Sci. 2020, 21, 2352. [Google Scholar] [CrossRef] [Green Version]
- Ma, D.; Hou, L.; Xia, H.; Li, H.; Fan, H.; Jia, X.; Niu, Z. PER2 inhibits proliferation and stemness of glioma stem cells via the Wnt/β-catenin signaling pathway. Oncol. Rep. 2020, 44, 533–542. [Google Scholar] [CrossRef]
- Zhang, Y.; Devocelle, A.; Souza, L.; Foudi, A.; Bento, S.T.; Desterke, C.; Sherrard, R.; Ballesta, A.; Adam, R.; Giron-Michel, J.; et al. BMAL1 knockdown triggers different colon carcinoma cell fates by altering the delicate equilibrium between AKT/mTOR and P53/P21 pathways. Aging (Albany NY) 2020, 12, 8067–8083. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wood, P.A.; Ansell, C.M.; Ohmori, M.; Oh, E.Y.; Xiong, Y.; Berger, F.G.; Peña, M.M.; Hrushesky, W.J. Beta-catenin induces beta-TrCP-mediated PER2 degradation altering circadian clock gene expression in intestinal mucosa of ApcMin/+ mice. J. Biochem. 2009, 145, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Damato, A.R.; Herzog, E.D. Circadian clock synchrony and chronotherapy opportunities in cancer treatment. Semin. Cell Dev. Biol. 2022, 126, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Maringe, C.; Spicer, J.; Morris, M.; Purushotham, A.; Nolte, E.; Sullivan, R.; Rachet, B.; Aggarwal, A. The impact of the COVID-19 pandemic on cancer deaths due to delays in diagnosis in England, UK: A national, population-based, modelling study. Lancet Oncol. 2020, 21, 1023–1034. [Google Scholar] [CrossRef]
- Fu, L.; Kettner, N.M. The circadian clock in cancer development and therapy. Prog. Mol. Biol. Transl. Sci. 2013, 119, 221–282. [Google Scholar] [CrossRef] [Green Version]
- Hasakova, K.; Vician, M.; Reis, R.; Zeman, M.; Herichova, I. The expression of clock genes cry1 and cry2 in human colorectal cancer and tumor adjacent tissues correlates differently dependent on tumor location. Neoplasma 2018, 65, 986–992. [Google Scholar] [CrossRef]
- Zeman, M.; Vician, M.; Monosíková, J.; Reis, R.; Herichová, I. Deregulated expression of the per2 gene in human colorectal carcinoma. Mol. Med. Rep. 2008, 1, 599–603. [Google Scholar] [CrossRef]
- Soták, M.; Polidarová, L.; Ergang, P.; Sumová, A.; Pácha, J. An association between clock genes and clock-controlled cell cycle genes in murine colorectal tumors. Int. J. Cancer 2013, 132, 1032–1041. [Google Scholar] [CrossRef]
- Hasakova, K.; Reis, R.; Vician, M.; Zeman, M.; Herichova, I. Expression of miR-34a-5p is up-regulated in human colorectal cancer and correlates with survival and clock gene PER2 expression. PLoS ONE 2019, 14, e0224396. [Google Scholar] [CrossRef] [Green Version]
- Misso, G.; Di Martino, M.T.; De Rosa, G.; Farooqi, A.A.; Lombardi, A.; Campani, V.; Zarone, M.R.; Gullà, A.; Tagliaferri, P.; Tassone, P.; et al. Mir-34: A new weapon against cancer? Mol. Ther. Nucleic Acids 2014, 3, e194. [Google Scholar] [CrossRef] [PubMed]
- Goblirsch, M.; Richtig, G.; Slaby, O.; Berindan-Neagoe, I.; Gerger, A.; Pichler, M. MicroRNAs as a tool to aid stratification of colorectal cancer patients and to guide therapy. Pharmacogenomics 2017, 18, 1027–1038. [Google Scholar] [CrossRef]
- Slaby, O.; Calin, G.A. Non-coding RNAs in Colorectal Cancer. Adv. Exp. Med. Biol. 2016, 937, 1–251. [Google Scholar] [CrossRef]
- Machackova, T.; Prochazka, V.; Kala, Z.; Slaby, O. Translational Potential of MicroRNAs for Preoperative Staging and Prediction of Chemoradiotherapy Response in Rectal Cancer. Cancers 2019, 11, 1545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Liao, Y.; Tang, L. MicroRNA-34 family: A potential tumor suppressor and therapeutic candidate in cancer. J. Exp. Clin. Cancer Res. 2019, 38, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. elife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Dweep, H.; Gretz, N. MiRWalk2.0: A comprehensive atlas of microRNA-target interactions. Nat. Methods 2015, 12, 697. [Google Scholar] [CrossRef]
- Lu, S.; Mukkada, V.A.; Mangray, S.; Cleveland, K.; Shillingford, N.; Schorl, C.; Brodsky, A.S.; Resnick, M.B. MicroRNA profiling in mucosal biopsies of eosinophilic esophagitis patients pre and post treatment with steroids and relationship with mRNA targets. PLoS ONE 2012, 7, e40676. [Google Scholar] [CrossRef]
- Place, R.F.; Li, L.C.; Pookot, D.; Noonan, E.J.; Dahiya, R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl. Acad. Sci. USA 2008, 105, 1608–1613. [Google Scholar] [CrossRef] [Green Version]
- Sadakierska-Chudy, A. MicroRNAs: Diverse Mechanisms of Action and Their Potential Applications as Cancer Epi-Therapeutics. Biomolecules 2020, 10, 1285. [Google Scholar] [CrossRef]
- Dreos, R.; Ambrosini, G.; Périer, R.C.; Bucher, P. The Eukaryotic Promoter Database: Expansion of EPDnew and new promoter analysis tools. Nucleic Acids Res. 2015, 43, D92–D96. [Google Scholar] [CrossRef] [PubMed]
- Yamakuchi, M.; Ferlito, M.; Lowenstein, C.J. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13421–13426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, F.; Yang, M.; Chen, Y.; Chen, W.; Wang, W. miR-34a induces immunosuppression in colorectal carcinoma through modulating a SIRT1/NF-κB/B7-H3/TNF-α axis. Cancer Immunol. Immunother. 2021, 70, 2247–2259. [Google Scholar] [CrossRef] [PubMed]
- Akao, Y.; Noguchi, S.; Iio, A.; Kojima, K.; Takagi, T.; Naoe, T. Dysregulation of microRNA-34a expression causes drug-resistance to 5-FU in human colon cancer DLD-1 cells. Cancer Lett. 2011, 300, 197–204. [Google Scholar] [CrossRef]
- Lai, M.; Du, G.; Shi, R.; Yao, J.; Yang, G.; Wei, Y.; Zhang, D.; Xu, Z.; Zhang, R.; Li, Y.; et al. MiR-34a inhibits migration and invasion by regulating the SIRT1/p53 pathway in human SW480 cells. Mol. Med. Rep. 2015, 11, 3301–3307. [Google Scholar] [CrossRef] [Green Version]
- Mohan, M.; Kumar, V.; Lackner, A.A.; Alvarez, X. Dysregulated miR-34a-SIRT1-acetyl p65 axis is a potential mediator of immune activation in the colon during chronic simian immunodeficiency virus infection of rhesus macaques. J. Immunol. 2015, 194, 291–306. [Google Scholar] [CrossRef] [Green Version]
- Martini, S.; Zuco, V.; Tortoreto, M.; Percio, S.; Campi, E.; Bezawy, R.E.; Doldi, V.; Landesman, Y.; Pennati, M.; Zaffaroni, N. Mir-34a-mediated survivin inhibition improves the antitumor activity of selinexor in triple-negative breast cancer. Pharmaceuticals 2021, 14, 523. [Google Scholar] [CrossRef]
- Cao, W.; Yang, W.; Fan, R.; Li, H.; Jiang, J.; Geng, M.; Jin, Y.; Wu, Y. miR-34a regulates cisplatin-induce gastric cancer cell death by modulating PI3K/AKT/survivin pathway. Tumor Biol. 2014, 35, 1287–1295. [Google Scholar] [CrossRef]
- Yang, B.; Huang, J.; Liu, H.; Guo, W.; Li, G. miR-335 directly, while miR-34a indirectly modulate survivin expression and regulate growth, apoptosis, and invasion of gastric cancer cells. Tumour. Biol. 2016, 37, 1771–1779. [Google Scholar] [CrossRef]
- Peng, Y.; Fan, J.Y.; Xiong, J.; Lo, Y.; Zhu, Y. Mir-34a enhances the susceptibility of gastric cancer to platycodin d by targeting surviving. Pathobiology 2019, 86, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Li, N.; Dong, Y.; Li, S.; Xu, L.; Li, X.; Li, Y.; Li, Z.; Ng, S.S.; Sung, J.J.; et al. miR-34a-5p suppresses colorectal cancer metastasis and predicts recurrence in patients with stage II/III colorectal cancer. Oncogene 2015, 34, 4142–4152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondratov, R.V.; Shamanna, R.K.; Kondratova, A.A.; Gorbacheva, V.Y.; Antoch, M.P. Dual role of the CLOCK/BMAL1 circadian complex in transcriptional regulation. FASEB J. 2006, 20, 530–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondratov, R.V.; Kondratova, A.A.; Lee, C.; Gorbacheva, V.Y.; Chernov, M.V.; Antoch, M.P. Post-translational regulation of circadian transcriptional CLOCK(NPAS2)/BMAL1 complex by CRYPTOCHROMES. Cell Cycle 2006, 5, 890–895. [Google Scholar] [CrossRef]
- Oda, A.; Katayose, Y.; Yabuuchi, S.; Yamamoto, K.; Mizuma, M.; Shirasou, S.; Onogawa, T.; Ohtsuka, H.; Yoshida, H.; Hayashi, H.; et al. Clock gene mouse period2 overexpression inhibits growth of human pancreatic cancer cells and has synergistic effect with cisplatin. Anticancer Res. 2009, 29, 1201–1209. [Google Scholar] [PubMed]
- Chiou, Y.Y.; Yang, Y.; Rashid, N.; Ye, R.; Selby, C.P.; Sancar, A. Mammalian Period represses and de-represses transcription by displacing CLOCK-BMAL1 from promoters in a Cryptochrome-dependent manner. Proc. Natl. Acad. Sci. USA 2016, 113, E6072–E6079. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Gustafson, C.L.; Sammons, P.J.; Khan, S.K.; Parsley, N.C.; Ramanathan, C.; Lee, H.W.; Liu, A.C.; Partch, C.L. Cryptochrome 1 regulates the circadian clock through dynamic interactions with the BMAL1 C terminus. Nat. Struct. Mol. Biol. 2015, 22, 476–484. [Google Scholar] [CrossRef] [Green Version]
- van der Horst, G.T.; Muijtjens, M.; Kobayashi, K.; Takano, R.; Kanno, S.; Takao, M.; de Wit, J.; Verkerk, A.; Eker, A.P.; van Leenen, D.; et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 1999, 398, 627–630. [Google Scholar] [CrossRef]
- Vitaterna, M.H.; Selby, C.P.; Todo, T.; Niwa, H.; Thompson, C.; Fruechte, E.M.; Hitomi, K.; Thresher, R.J.; Ishikawa, T.; Miyazaki, J.; et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc. Natl. Acad. Sci. USA 1999, 96, 12114–12119. [Google Scholar] [CrossRef]
- Selby, C.P.; Thompson, C.; Schmitz, T.M.; Van Gelder, R.N.; Sancar, A. Functional redundancy of cryptochromes and classical photoreceptors for nonvisual ocularphotoreception in mice. Proc. Natl. Acad. Sci. USA 2000, 97, 14697–14702. [Google Scholar] [CrossRef] [Green Version]
- Karki, N.; Vergish, S.; Zoltowski, B.D. Cryptochromes: Photochemical and structural insight into magnetoreception. Protein Sci. 2021, 30, 1521–1534. [Google Scholar] [CrossRef] [PubMed]
- Kyriacou, C.P.; Rosato, E. Genetic analysis of cryptochrome in insect magnetosensitivity. Front. Physiol. 2022, 13, 928416. [Google Scholar] [CrossRef] [PubMed]
- Yoshii, T.; Ahmad, M.; Helfrich-Förster, C. Cryptochrome mediates light-dependent magnetosensitivity of Drosophila’s circadian clock. PLoS Biol. 2009, 7, e1000086. [Google Scholar] [CrossRef]
- Fedele, G.; Edwards, M.D.; Bhutani, S.; Hares, J.M.; Murbach, M.; Green, E.W.; Dissel, S.; Hastings, M.H.; Rosato, E.; Kyriacou, C.P. Genetic analysis of circadian responses to low frequency electromagnetic fields in Drosophila melanogaster. PLoS Genet. 2014, 10, e1004804. [Google Scholar] [CrossRef] [Green Version]
- Foley, L.E.; Gegear, R.J.; Reppert, S.M. Human cryptochrome exhibits light-dependent magnetosensitivity. Nat. Commun. 2011, 2, 356. [Google Scholar] [CrossRef] [Green Version]
- Vieira, J.; Jones, A.R.; Danon, A.; Sakuma, M.; Hoang, N.; Robles, D.; Tait, S.; Heyes, D.J.; Picot, M.; Yoshii, T.; et al. Human cryptochrome-1 confers light independent biological activity in transgenic Drosophila correlated with flavin radical stability. PLoS ONE 2012, 7, e31867. [Google Scholar] [CrossRef] [Green Version]
- Sherrard, R.M.; Morellini, N.; Jourdan, N.; El-Esawi, M.; Arthaut, L.D.; Niessner, C.; Rouyer, F.; Klarsfeld, A.; Doulazmi, M.; Witczak, J.; et al. Low-intensity electromagnetic fields induce human cryptochrome to modulate intracellular reactive oxygen species. PLoS Biol. 2018, 16, e2006229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pooam, M.; Jourdan, N.; Aguida, B.; Dahon, C.; Baouz, S.; Terry, C.; Raad, H.; Ahmad, M. Exposure to 1.8 GHz radiofrequency field modulates ROS in human HEK293 cells as a function of signal amplitude. Commun. Integr. Biol. 2022, 15, 54–66. [Google Scholar] [CrossRef]
- Zeng, Z.; Wei, J.; Liu, Y.; Zhang, W.; Mabe, T. Magnetoreception of Photoactivated Cryptochrome 1 in Electrochemistry and Electron Transfer. ACS Omega 2018, 3, 4752–4759. [Google Scholar] [CrossRef]
- Granger, J.; Cummer, S.A.; Lohmann, K.J.; Johnsen, S. Environmental sources of radio frequency noise: Potential impacts on magnetoreception. J. Comp. Physiol. A Neuroethol. Sens. Neural. Behav. Physiol. 2022, 208, 83–95. [Google Scholar] [CrossRef]
- Pizarro, A.; Hayer, K.; Lahens, N.F.; Hogenesch, J.B. CircaDB: A database of mammalian circadian gene expression profiles. Nucleic Acids Res. 2013, 41, D1009–D1013. [Google Scholar] [CrossRef] [PubMed]
- Storcelová, M.; Vicián, M.; Reis, R.; Zeman, M.; Herichová, I. Expression of cell cycle regulatory factors hus1, gadd45a, rb1, cdkn2a and mre11a correlates with expression of clock gene per2 in human colorectal carcinoma tissue. Mol. Biol. Rep. 2013, 40, 6351–6361. [Google Scholar] [CrossRef] [PubMed]
- Lei, K.F.; Wang, Y.H.; Chen, H.Y.; Sun, J.H.; Cheng, J.Y. Electrokinetic acceleration of DNA hybridization in microsystems. Talanta 2015, 138, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Erdal, M.E.; Yılmaz, S.G.; Gürgül, S.; Uzun, C.; Derici, D.; Erdal, N. miRNA expression profile is altered differentially in the rat brain compared to blood after experimental exposure to 50 Hz and 1 mT electromagnetic field. Prog. Biophys. Mol. Biol. 2018, 132, 35–42. [Google Scholar] [CrossRef]
- Dasdag, S.; Akdag, M.Z.; Erdal, M.E.; Erdal, N.; Ay, O.I.; Ay, M.E.; Yilmaz, S.G.; Tasdelen, B.; Yegin, K. Long term and excessive use of 900 MHz radiofrequency radiation alter microRNA expression in brain. Int. J. Radiat. Biol. 2015, 91, 306–311. [Google Scholar] [CrossRef]
- Dasdag, S.; Akdag, M.Z.; Erdal, M.E.; Erdal, N.; Ay, O.I.; Ay, M.E.; Yilmaz, S.G.; Tasdelen, B.; Yegin, K. Effects of 2.4 GHz radiofrequency radiation emitted from Wi-Fi equipment on microRNA expression in brain tissue. Int. J. Radiat. Biol. 2015, 91, 555–561. [Google Scholar] [CrossRef]
- Dasdag, S.; Akdag, M.Z.; Bashan, M.; Kizmaz, V.; Erdal, N.; Emin Erdal, M.; Tughan Kiziltug, M.; Yegin, K. Role of 2.4 GHz radiofrequency radiation emitted from Wi-Fi on some miRNA and faty acids composition in brain. Electromagn. Biol. Med. 2022, 41, 281–292. [Google Scholar] [CrossRef]
- Sun, M.; Du, M.; Zhang, W.; Xiong, S.; Gong, X.; Lei, P.; Zha, J.; Zhu, H.; Li, H.; Huang, D.; et al. Survival and Clinicopathological Significance of SIRT1 Expression in Cancers: A Meta-Analysis. Front. Endocrinol. (Lausanne) 2019, 10, 121. [Google Scholar] [CrossRef]
- Cheung, C.H.; Huang, C.C.; Tsai, F.Y.; Lee, J.Y.; Cheng, S.M.; Chang, Y.C.; Huang, Y.C.; Chen, S.H.; Chang, J.Y. Survivin- biology and potential as a therapeutic target in oncology. OncoTargets Ther. 2013, 6, 1453–1462. [Google Scholar] [CrossRef]
- Yu, H.; Meng, X.; Wu, J.; Pan, C.; Ying, X.; Zhou, Y.; Liu, R.; Huang, W. Cryptochrome 1 overexpression correlates with tumor progression and poor prognosis in patients with colorectal cancer. PLoS ONE 2013, 8, e61679. [Google Scholar] [CrossRef] [Green Version]
- Aroca-Siendones, M.I.; Moreno-SanJuan, S.; Puentes-Pardo, J.D.; Verbeni, M.; Arnedo, J.; Escudero-Feliu, J.; García-Costela, M.; García-Robles, A.; Carazo, Á.; León, J. Core Circadian Clock Proteins as Biomarkers of Progression in Colorectal Cancer. Biomedicines 2021, 9, 967. [Google Scholar] [CrossRef] [PubMed]
- Mazzoccoli, G.; Colangelo, T.; Panza, A.; Rubino, R.; De Cata, A.; Tiberio, C.; Valvano, M.R.; Pazienza, V.; Merla, G.; Augello, B.; et al. Deregulated expression of cryptochrome genes in human colorectal cancer. Mol. Cancer 2016, 15, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, L.; Yu, Y.; Sun, S.; Zhang, T.; Wang, M. Cry 1 Regulates the Clock Gene Network and Promotes Proliferation and Migration Via the Akt/P53/P21 Pathway in Human Osteosarcoma Cells. J. Cancer 2018, 9, 2480–2491. [Google Scholar] [CrossRef]
- Liu, H.; Liu, Y.; Hai, R.; Liao, W.; Luo, X. The role of circadian clocks in cancer: Mechanisms and clinical implications. Genes Dis. 2022, in press. [Google Scholar] [CrossRef]
- Fang, L.; Yang, Z.; Zhou, J.; Tung, J.Y.; Hsiao, C.D.; Wang, L.; Deng, Y.; Wang, P.; Wang, J.; Lee, M.H. Circadian Clock Gene CRY2 Degradation Is Involved in Chemoresistance of Colorectal Cancer. Mol. Cancer Ther. 2015, 14, 1476–1487. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Z.L.; Wu, M.W.; Sun, J.; Sun, Y.L.; Cai, Y.C.; Huang, Y.J.; Xian, L.J. Effects of the biological clock gene Bmal1 on tumour growth and anti-cancer drug activity. J. Biochem. 2010, 148, 319–326. [Google Scholar] [CrossRef]
- Fuhr, L.; El-Athman, R.; Scrima, R.; Cela, O.; Carbone, A.; Knoop, H.; Li, Y.; Hoffmann, K.; Laukkanen, M.O.; Corcione, F.; et al. The Circadian Clock Regulates Metabolic Phenotype Rewiring Via HKDC1 and Modulates Tumor Progression and Drug Response in Colorectal Cancer. eBioMedicine 2018, 33, 105–121. [Google Scholar] [CrossRef] [Green Version]
- Wood, P.A.; Yang, X.; Hrushesky, W.J.M. The Role of Circadian Rhythm in the Pathogenesis of Colorectal Cancer. Curr. Colorectal. Cancer Rep. 2010, 6, 74–82. [Google Scholar] [CrossRef]
- Fu, L.; Pelicano, H.; Liu, J.; Huang, P.; Lee, C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 2002, 111, 41–50. [Google Scholar] [CrossRef]
- Yang, X.; He, X.; Yang, Z.; Jabbari, E. Mammalian PER2 regulates AKT activation and DNA damage response. Biochem. Cell Biol. 2012, 90, 675–682. [Google Scholar] [CrossRef] [Green Version]
- Wood, P.A.; Yang, X.; Taber, A.; Oh, E.Y.; Ansell, C.; Ayers, S.E.; Al-Assaad, Z.; Carnevale, K.; Berger, F.G.; Peña, M.M.; et al. Period 2 mutation accelerates ApcMin/+ tumorigenesis. Mol. Cancer Res. 2008, 6, 1786–1793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Gong, X.; Yang, K. Overexpression of the clock gene Per2 suppresses oral squamous cell carcinoma progression by activating autophagy via the PI3K/AKT/mTOR pathway. J. Cancer 2020, 11, 3655–3666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Ao, Y.; Yang, K.; Tang, H.; Chen, D. Circadian clock gene Per2 plays an important role in cell proliferation, apoptosis and cell cycle progression in human oral squamous cell carcinoma. Oncol. Rep. 2016, 35, 3387–3394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazzoccoli, G.; Panza, A.; Valvano, M.R.; Palumbo, O.; Carella, M.; Pazienza, V.; Biscaglia, G.; Tavano, F.; Di Sebastiano, P.; Andriulli, A.; et al. Clock gene expression levels and relationship with clinical and pathological features in colorectal cancer patients. Chronobiol. Int. 2011, 28, 841–851. [Google Scholar] [CrossRef]
- Mehta, N.; Cheng, H.Y. Micro-managing the circadian clock: The role of microRNAs in biological timekeeping. J. Mol. Biol. 2013, 425, 3609–3624. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, J.; Li, N.; Liu, Z.; Chen, Z.; Li, Z.; Lai, Y.; Shen, L.; Gao, J. miR-34a increases the sensitivity of colorectal cancer cells to 5-fluorouracil in vitro and in vivo. Am. J. Cancer Res. 2018, 8, 280–290. [Google Scholar]
- Lu, H.; Hao, L.; Yang, H.; Chen, J.; Liu, J. miRNA-34a suppresses colon carcinoma proliferation and induces cell apoptosis by targeting SYT1. Int. J. Clin. Exp. Pathol. 2019, 12, 2887–2897. [Google Scholar]
- Hehlgans, S.; Petraki, C.; Reichert, S.; Cordes, N.; Rödel, C.; Rödel, F. Double targeting of Survivin and XIAP radiosensitizes 3D grown human colorectal tumor cells and decreases migration. Radiother. Oncol. 2013, 108, 32–39. [Google Scholar] [CrossRef]
- George, R.; Hehlgans, S.; Fleischmann, M.; Rödel, C.; Fokas, E.; Rödel, F. Advances in nanotechnology-based platforms for survivin-targeted drug discovery. Expert Opin. Drug Discov. 2022, 17, 733–754. [Google Scholar] [CrossRef]
- Halgamuge, M.N.; Skafidas, E.; Davis, D. A meta-analysis of in vitro exposures to weak radiofrequency radiation exposure from mobile phones (1990–2015). Environ. Res. 2020, 184, 109227. [Google Scholar] [CrossRef]
- Šimaiová, V.; Almášiová, V.; Holovská, K.; Kisková, T.; Horváthová, F.; Ševčíková, Z.; Tóth, Š.; Raček, A.; Račeková, E.; Beňová, K.; et al. The effect of 2.45 GHz non-ionizing radiation on the structure and ultrastructure of the testis in juvenile rats. Histol. Histopathol. 2019, 34, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Holovská, K.; Almášiová, V.; Cigánková, V.; Beňová, K.; Račeková, E.; Martončíková, M. Structural and ultrastructural study of rat liver influenced by electromagnetic radiation. J. Toxicol. Environ. Health A 2015, 78, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Raček, A.; Beňová, K.; Arnoul, P.; Závodská, M.; Angelidis, A.; Cigánková, V.; Šimaiová, V.; Račeková, E. Age-dependent effect of long-term microwave radiation on postnatal neurogenesis in rats: Morphological and behavioral study. Physiol. Res. 2018, 67, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Ozgur, E.; Kayhan, H.; Kismali, G.; Senturk, F.; Sensoz, M.; Ozturk, G.G.; Sel, T. Effects of radiofrequency radiation on colorectal cancer cell proliferation and inflammation. Turk. J. Biochem. 2021, 46, 525–532. [Google Scholar] [CrossRef]
- Gökçen, S.; Kurt, B.; Küçükbağrıaçık, Y.; Ozgur-Buyukatalay, E.; Kismali, G. Effects of radiofrequency radiation on apoptotic and antiapoptotic factors in colorectal cancer cells. Electromagn. Biol. Med. 2022, 41, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Obernosterer, G.; Leuschner, P.J.; Alenius, M.; Martinez, J. Post-transcriptional regulation of microRNA expression. RNA 2006, 12, 1161–1167. [Google Scholar] [CrossRef] [Green Version]
- Graves, P.; Zeng, Y. Biogenesis of mammalian microRNAs: A global view. Genom. Proteom. Bioinform. 2012, 10, 239–245. [Google Scholar] [CrossRef] [Green Version]
- Herichová, I.; Tesáková, B.; Kršková, L.; Olexová, L. Food reward induction of rhythmic clock gene expression in the prefrontal cortex of rats is accompanied by changes in miR-34a-5p expression. Eur. J. Neurosci. 2021, 54, 7476–7492. [Google Scholar] [CrossRef]
- Jafari, N.; Abediankenari, S.; Hossein-Nataj, H. miR-34a mimic or pre-mir-34a, which is the better option for cancer therapy? KatoIII as a model to study miRNA action in human gastric cancer cells. Cancer Cell Int. 2021, 21, 178. [Google Scholar] [CrossRef]
- Liu, G.; Min, H.; Yue, S.; Chen, C.Z. Pre-miRNA loop nucleotides control the distinct activities of mir-181a-1 and mir-181c in early T cell development. PLoS ONE 2008, 3, e3592. [Google Scholar] [CrossRef] [Green Version]
- Momin, M.Y.; Gaddam, R.R.; Kravitz, M.; Gupta, A.; Vikram, A. The Challenges and Opportunities in the Development of MicroRNA Therapeutics: A Multidisciplinary Viewpoint. Cells 2021, 10, 3097. [Google Scholar] [CrossRef] [PubMed]










Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olejárová, S.; Moravčík, R.; Herichová, I. 2.4 GHz Electromagnetic Field Influences the Response of the Circadian Oscillator in the Colorectal Cancer Cell Line DLD1 to miR-34a-Mediated Regulation. Int. J. Mol. Sci. 2022, 23, 13210. https://doi.org/10.3390/ijms232113210
Olejárová S, Moravčík R, Herichová I. 2.4 GHz Electromagnetic Field Influences the Response of the Circadian Oscillator in the Colorectal Cancer Cell Line DLD1 to miR-34a-Mediated Regulation. International Journal of Molecular Sciences. 2022; 23(21):13210. https://doi.org/10.3390/ijms232113210
Chicago/Turabian StyleOlejárová, Soňa, Roman Moravčík, and Iveta Herichová. 2022. "2.4 GHz Electromagnetic Field Influences the Response of the Circadian Oscillator in the Colorectal Cancer Cell Line DLD1 to miR-34a-Mediated Regulation" International Journal of Molecular Sciences 23, no. 21: 13210. https://doi.org/10.3390/ijms232113210
APA StyleOlejárová, S., Moravčík, R., & Herichová, I. (2022). 2.4 GHz Electromagnetic Field Influences the Response of the Circadian Oscillator in the Colorectal Cancer Cell Line DLD1 to miR-34a-Mediated Regulation. International Journal of Molecular Sciences, 23(21), 13210. https://doi.org/10.3390/ijms232113210