Metabolic Activation and DNA Interactions of Carcinogenic N-Nitrosamines to Which Humans Are Commonly Exposed
Abstract
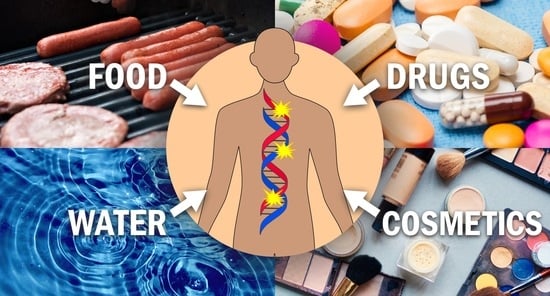
:1. Introduction
2. Overview of Carcinogenic N-Nitrosamines to Which Humans Are Commonly Exposed
2.1. Carcinogenic N-Nitrosamines Occurring in Food
2.2. Carcinogenic N-Nitrosamines Occurring in Water
2.3. Carcinogenic N-Nitrosamines Occurring in Drugs
2.4. Carcinogenic N-Nitrosamines Occurring in Cosmetics
3. Metabolic Activation and DNA Interactions of Carcinogenic Acyclic N-Nitrosamines
3.1. N-Nitrosodimethylamine (NDMA)
3.1.1. Exposure and Carcinogenicity
3.1.2. Metabolism
3.1.3. Methyl DNA Adducts Formed by NDMA Metabolism
3.1.4. Mutagenicity and Genotoxicity of Methyl DNA Adducts
3.1.5. Methyl DNA Adducts in Human Tissues
3.2. N-Nitrosomethylethylamine (NMEA)
3.2.1. Exposure and Carcinogenicity
3.2.2. Metabolism
3.2.3. DNA Adducts Formed by NMEA Metabolism
3.3. N-Nitrososarcosine (NSAR)
3.3.1. Exposure and Carcinogenicity
3.3.2. Metabolism
3.3.3. Carboxymethylating and Methylating Intermediates Formed by NSAR Metabolism
3.3.4. Mutagenicity and Genotoxicity of Carboxymethyl DNA Adducts
3.3.5. Carboxymethyl DNA Adducts in Human Tissues
3.4. N-Nitrosodiethylamine (NDEA)
3.4.1. Exposure and Carcinogenicity
3.4.2. Metabolism
3.4.3. Ethyl DNA Adducts Formed by NDEA Metabolism
3.4.4. Mutagenicity and Genotoxicity of Ethyl DNA Adducts
3.4.5. Ethyl DNA Adducts in Human Tissues
3.5. N-Nitroso-di-n-propylamine (NDPA)
3.5.1. Exposure and Carcinogenicity
3.5.2. Metabolism
3.5.3. DNA Adducts Formed by NDPA Metabolism
3.6. N-Nitrosodiethanolamine (NDELA)
3.6.1. Exposure and Carcinogenicity
3.6.2. Metabolism
3.6.3. DNA Adducts Formed by NDELA Metabolism
3.7. N-Nitrosodi-n-butylamine (NDBA)
3.7.1. Exposure and Carcinogenicity
3.7.2. Metabolism
3.7.3. DNA Adducts Formed by NDBA Metabolism
4. Metabolic Activation and DNA Interactions of Carcinogenic Cyclic N-Nitrosamines
4.1. N-Nitrosopyrrolidine (NPYR)
4.1.1. Exposure and Carcinogenicity
4.1.2. Metabolism
4.1.3. DNA Adducts Formed by NPYR Metabolism
DNA Adducts Formed by NPYR-Derived Carbocations
DNA Adducts Formed by an NPYR-Derived Oxonium Ion
DNA Adducts Formed by NPYR-Derived Crotonaldehyde
Other DNA Adducts Related to NPYR Metabolism
4.1.4. Mutagenicity and Genotoxicity of NPYR-Derived DNA Adducts
4.1.5. Human DNA Adducts Related to NPYR Metabolism
4.2. N-Nitrosopiperidine (NPIP)
4.2.1. Exposure and Carcinogenicity
4.2.2. Metabolism
4.2.3. DNA Adducts Formed by NPIP Metabolism
4.2.4. Human DNA Adducts Related to NPIP Metabolism
4.3. N-Nitrosomorpholine (NMOR)
4.3.1. Exposure and Carcinogenicity
4.3.2. Metabolism
4.3.3. DNA Adducts Formed by NMOR Metabolism
5. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| α-AcetoxyNPYR | α-acetoxy-N-nitrosopyrrolidine |
| AGT | O6-alkylguanine-DNA alkyltransferease |
| APNG | N-(N′-acetyl-L-prolyl)-N-nitrosoglycine |
| AS | azaserine |
| BBN | N-nitroso-(4-hydroxybutyl)butylamine |
| E. coli | Escherichia coli |
| FDA | Food and Drug Administration |
| GSH | glutathione |
| α-hydroxyNMOR | N-nitroso-3-hydroxymorpholine |
| IARC | International Agency for Research on Cancer |
| i.p. | intraperitoneal |
| i.v. | intravenous |
| KDA | potassium diazoacetate |
| MGMT | O6-methylguanine-DNA methyltransferase |
| MNPA | 3-(methylnitrosamino)propionic acid |
| N2-THF-dGuo | N2-(tetrahydrofuran-2-yl)-2′-deoxyguanosine |
| N2-(4-HOB)-dGuo | N2-(4-hydroxybutyl)-2′-deoxyguanosine |
| N7-CE-Gua | N7-(2′-carboxyethyl)guanine |
| N7-CM-Gua | N7-(carboxymethyl)guanine |
| N7-Et-Gua | N7-ethylguanine |
| N7-Me-Gua | N7-methylguanine |
| N7-n-Pr-Gua | N7-(n-propyl)guanine |
| N7,8-butano-Gua | 2-amino-6,7,8,9-tetrahydro-9-hydroxypyrido [2,1-f]purine-4(3H)-one |
| N7,8-Cro-Gua | 2-amino-7,8-dihydro-8-hydroxy-6-methyl-3H-pyrrolo [2,1-f]purine-4(6H)-one |
| NDBA | N-nitrosodi-n-butylamine |
| NDEA | N-nitrosodiethylamine |
| NDELA | N-nitrosodiethanolamine |
| NDIPA | N-nitrodiisopropylamine |
| NDMA | N-nitrosodimethylamine |
| NDPA | N-nitrosodi-n-propylamine |
| NER | nucleotide excision repair |
| NHEG | N-nitroso-(2-hydroxyethyl)glycine |
| NHMOR | N-Nitroso-2-hydroxymorpholine |
| NHPPA | N-nitroso-2-hydroxypropylpropylamine |
| NIPEA | N-nitrosoisopropylethylamine |
| NMBA | N-nitroso-N-methyl-4-aminobutanoic acid |
| NMEA | N-nitrosomethylethylamine |
| NMOR | N-nitrosomorpholine |
| NMPA | N-nitrosomethylphenylamine |
| NOPPA | N-nitroso-2-oxopropylpropylamine |
| NPIP | N-nitrosopiperidine |
| NPYR | N-nitrosopyrrolidine |
| NSAR | N-nitrososarcosine |
| 7-(2-oxopropyl)-N1,N2-etheno-dGuo | 7-(2-oxopropyl)-5,9-dihydro-9-oxo-3-β-D-deoxyribofuranosylimidazo [1,2-a]purine |
| O2-Me-Thd | O2-methylthymidine |
| O4-Me-Thd | O4-methylthymidine |
| O6-n-Bu-Gua | O6-(n-butyl)guanine |
| O6-CM-Gua | O6-(carboxymethyl)guanine |
| O6-Et-Gua | O6-ethylguanine |
| O6-(4-OH-n-Bu)-Gua | O6-(4-hydroxybutyl)guanine |
| O6-HOEt-dGuo | O6-(2-hydroxyethyl)-2′-deoxyguanosine |
| O6-Me-Gua | O6-methylguanine |
| pol κ | polymerase κ |
| ppm | parts per million |
| s.c. | subcutaneous |
| S. typhinurium | Salmonella typhinurium |
| THP-OH | 2-hydroxytetrahydropyran |
References
- Magee, P.N.; Barnes, J.M. The production of malignant primary hepatic tumours in the rat by feeding dimethylnitrosamine. Br. J. Cancer 1956, 10, 114–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakshaug, J.; Sognen, E.; Hansen, M.A.; Koppang, N. Dimethylnitrosamine; its hepatotoxic effect in sheep and its occurrence in toxic batches of herring meal. Nature 1965, 206, 1261–1262. [Google Scholar] [CrossRef] [PubMed]
- Ender, F.; Ceh, L. Occurrence of nitrosamines in foodstuffs for human and animal consumption. Food Cosmet. Toxicol. 1968, 6, 569–571. [Google Scholar] [CrossRef]
- Sen, N.P. The evidence for the presence of dimethylnitrosamine in meat products. Food Cosmet. Toxicol. 1972, 10, 219–223. [Google Scholar] [CrossRef]
- Lijinsky, W. Chemistry and Biology of N-Nitroso Compounds; Cambridge University Press: Cambridge, UK, 1992. [Google Scholar]
- Druckrey, H.; Preussmann, R.; Ivankovic, S.; Schmähl, D. Organotropic carcinogenic effects of 65 various N-nitroso- compounds on BD rats. Z. Krebsforsch. 1967, 69, 103–201. [Google Scholar] [CrossRef]
- Preussmann, R.; Stewart, B.W. N-Nitroso Carcinogens. In Chemical Carcinogens, 2nd ed.; ACS Monograph 182; Searle, C.E., Ed.; American Chemical Society: Washington, DC, USA, 1984; Volume 2, pp. 643–828. [Google Scholar]
- Bogovski, P.; Bogovski, S. Animal species in which N-nitroso compounds induce cancer. Int. J. Cancer 1981, 27, 471–474. [Google Scholar] [CrossRef]
- Gushgari, A.J.; Halden, R.U. Critical review of major sources of human exposure to N-nitrosamines. Chemosphere 2018, 210, 1124–1136. [Google Scholar] [CrossRef]
- Kroes, R.; Kleiner, J.; Renwick, A. The threshold of toxicological concern concept in risk assessment. Toxicol. Sci. 2005, 86, 226–230. [Google Scholar] [CrossRef] [Green Version]
- Tricker, A.R. N-nitroso compounds and man: Sources of exposure, endogenous formation and occurrence in body fluids. Eur. J. Cancer Prev. 1997, 6, 226–268. [Google Scholar] [CrossRef]
- Park, J.E.; Seo, J.E.; Lee, J.Y.; Kwon, H. Distribution of seven N-nitrosamines in food. Toxicol. Res. 2015, 31, 279–288. [Google Scholar] [CrossRef] [Green Version]
- Mitch, W.A.; Sharp, J.O.; Trussell, R.R.; Valentine, R.L.; Alvarez-Cohen, L.; Sedlak, D.L. N-nitrosodimethylamine (NDMA) as a drinking water contaminant: A review. Environ. Eng. Sci. 2003, 20, 389–404. [Google Scholar] [CrossRef] [Green Version]
- Beard, J.C.; Swager, T.M. An organic chemist’s guide to N-nitrosamines: Their structure, reactivity, and role as contaminants. J. Org. Chem. 2021, 86, 2037–2057. [Google Scholar] [CrossRef] [PubMed]
- Bharate, S.S. Critical analysis of drug product recalls due to nitrosamine impurities. J. Med. Chem. 2021, 64, 2923–2936. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food and Drug Administration. Drug Recalls. Available online: https://www.fda.gov/drugs/drug-safety-and-availability/drug-recalls (accessed on 21 September 2021).
- U.S. Food and Drug Administration. Control of Nitrosamine Impurities in Human Drugs: Guidance for Industry; Center for Drug Evaluation and Research: Silver Spring, MD, USA, 2021.
- Lim, D.S.; Roh, T.H.; Kim, M.K.; Kwon, Y.C.; Choi, S.M.; Kwack, S.J.; Kim, K.B.; Yoon, S.; Kim, H.S.; Lee, B.M. Risk assessment of N-nitrosodiethylamine (NDEA) and N-nitrosodiethanolamine (NDELA) in cosmetics. J. Toxicol. Environ. Health A 2018, 81, 465–480. [Google Scholar] [CrossRef] [PubMed]
- Preussmann, R.; Spiegelhalder, B.; Eisenbrand, G.; Wurtele, G.; Hofmann, I. Urinary excretion of N-nitrosodiethanolamine in rats following its epicutaneous and intratracheal administration and its formation in vivo following skin application of diethanolamine. Cancer Lett. 1981, 13, 227–231. [Google Scholar] [CrossRef]
- American Chemical Society. Molecule of the Week Archive: N-Nitrosodimethylamine. ACS Molecule of the Week, 14 January 2019. [Google Scholar]
- Peto, R.; Gray, R.; Brantom, P.; Grasso, P. Effects on 4080 rats of chronic ingestion of N-nitrosodiethylamine or N-nitrosodimethylamine: A detailed dose-response study. Cancer Res. 1991, 51 Pt 2, 6415–6451. [Google Scholar]
- Peto, R.; Gray, R.; Brantom, P.; Grasso, P. Dose and time relationships for tumor induction in the liver and esophagus of 4080 inbred rats by chronic ingestion of N-nitrosodiethylamine or N-nitrosodimethylamine. Cancer Res. 1991, 51 Pt 2, 6452–6469. [Google Scholar]
- National Toxicology Program. Report on Carcinogens, 15th ed.; U.S. Department of Health and Human Services, Public Health Service: Research Triangle Park, NC, USA, 2021.
- Yamazaki, H.; Inui, Y.; Yun, C.H.; Guengerich, F.P.; Shimada, T. Cytochrome P450 2E1 and 2A6 enzymes as major catalysts for metabolic activation of N-nitrosodialkylamines and tobacco-related nitrosamines in human liver microsomes. Carcinogenesis 1992, 13, 1789–1794. [Google Scholar] [CrossRef]
- Mochizuki, M.; Osabe, M.; Anjo, T.; Suzuki, E.; Okada, M. Mutagenicity of α-hydroxy N-nitrosamines in V79 Chinese hamster cells. J. Cancer Res. Clin. Oncol. 1984, 108, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Chowdhury, G.; Calcutt, M.W.; Guengerich, F.P. Oxidation of N-nitrosoalkylamines by human cytochrome P450 2A6: Sequential oxidation to aldehydes and carboxylic acids and analysis of reaction steps. J. Biol. Chem. 2010, 285, 8031–8044. [Google Scholar] [CrossRef] [Green Version]
- Chowdhury, G.; Calcutt, M.W.; Nagy, L.D.; Guengerich, F.P. Oxidation of methyl and ethyl nitrosamines by cytochrome P450 2E1 and 2B1. Biochemistry 2012, 51, 9995–10007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Godoy, H.M.; Diaz Gomez, M.I.; Castro, J.A. Mechanism of dimethylnitrosamine metabolism and activation in rats. J. Natl. Cancer Inst. 1978, 61, 1285–1289. [Google Scholar] [CrossRef] [PubMed]
- Lake, B.G.; Minski, M.J.; Phillips, J.C.; Gangolli, S.D.; Lloyd, A.G. Investigations into the hepatic metabolism of dimethylnitrosamine in the rat. Life Sci. 1975, 17, 1599–1606. [Google Scholar] [CrossRef]
- Kroeger-Koepke, M.B.; Koepke, S.R.; McClusky, G.A.; Magee, P.N.; Michejda, C.J. α-Hydroxylation pathway in the in vitro metabolism of carcinogenic nitrosamines: N-nitrosodimethylamine and N-nitroso-N-methylaniline. Proc. Natl. Acad. Sci. USA 1981, 78, 6489–6493. [Google Scholar] [CrossRef] [Green Version]
- Cottrell, R.C.; Lake, B.G.; Phillips, J.C.; Gangolli, S.D. The hepatic metabolism of 15N labelled dimethylnitrosamine in the rat. Biochem. Pharmacol. 1977, 26, 809–813. [Google Scholar] [CrossRef]
- Koepke, S.R.; Tondeur, Y.; Farrelly, J.G.; Stewart, M.L.; Michejda, C.J.; Kroeger-Koepke, M.B. Metabolism of 15N-labelled N-nitrosodimethylamine and N-nitroso-N-methylaniline by isolated rat hepatocytes. Biochem. Pharmacol. 1984, 33, 1509–1513. [Google Scholar] [CrossRef]
- Swann, P.F.; Mace, R.; Angeles, R.M.; Keefer, L.K. Deuterium isotope effect on metabolism of N-nitrosodimethylamine in vivo in rat. Carcinogenesis 1983, 4, 821–825. [Google Scholar] [CrossRef]
- Umbenhauer, D.R.; Pegg, A.E. Alkylation of intracellular and extracellular DNA by dimethylnitrosamine following activation by isolated rat hepatocytes. Cancer Res. 1981, 41 Pt 1, 3471–3474. [Google Scholar]
- Yoo, J.S.; Ishizaki, H.; Yang, C.S. Roles of cytochrome P450IIE1 in the dealkylation and denitrosation of N-nitrosodimethylamine and N-nitrosodiethylamine in rat liver microsomes. Carcinogenesis 1990, 11, 2239–2243. [Google Scholar] [CrossRef]
- Lorr, N.A.; Tu, Y.Y.; Yang, C.S. The nature of nitrosamine denitrosation by rat liver microsomes. Carcinogenesis 1982, 3, 1039–1043. [Google Scholar] [CrossRef]
- Lee, V.M.; Keefer, L.K.; Archer, M.C. An evaluation of the roles of metabolic denitrosation and alpha-hydroxylation in the hepatotoxicity of N-Nitrosodimethylamine. Chem. Res. Toxicol. 1996, 9, 1319–1324. [Google Scholar] [CrossRef] [PubMed]
- Negishi, T.; Shiotani, T.; Fujikawa, K.; Hayatsu, H. The genotoxicities of N-nitrosamines in Drosophila melanogaster in vivo: The correlation of mutagenicity in the wing spot test with the DNA damages detected by the DNA-repair test. Mutat. Res. 1991, 252, 119–128. [Google Scholar] [CrossRef]
- Singer, B. Alkylation of the O6 of guanine is only one of many chemical events that may initiate carcinogenesis. Cancer Investig. 1984, 2, 233–238. [Google Scholar] [CrossRef]
- Li, Y.; Hecht, S.S. Metabolism and DNA adduct formation of tobacco-specific N-nitrosamines. Int. J. Mol. Sci. 2022, Manuscript submitted.
- Den Engelse, L.; Menkveld, G.J.; De Brij, R.J.; Tates, A.D. Formation and stability of alkylated pyrimidines and purines (including imidazole ring-opened 7-alkylguanine) and alkylphosphotriesters in liver DNA of adult rats treated with ethylnitrosourea or dimethylnitrosamine. Carcinogenesis 1986, 7, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Margison, G.P.; Margison, J.M.; Montesano, R. Methylated purines in the deoxyribonucleic acid of various Syrian-golden-hamster tissues after administration of a hepatocarcinogenic dose of dimethylnitrosamine. Biochem. J. 1976, 157, 627–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margison, G.P.; Margison, J.M.; Montesano, R. Accumulation of O6-methylguanine in non-target-tissue deoxyribonucleic acid during chronic administration of dimethylnitrosamine. Biochem. J. 1977, 165, 463–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hecht, S.S.; Trushin, N.; Castonguay, A.; Rivenson, A. Comparative tumorigenicity and DNA methylation in F344 rats by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and N-nitrosodimethylamine. Cancer Res. 1986, 46, 498–502. [Google Scholar]
- Montesano, R.; Brésil, H.; Margison, G.P. Increased excision of O6-methylguanine from rat liver DNA after chronic administration of dimethylnitrosamine. Cancer Res. 1979, 39, 1798–1802. [Google Scholar]
- Souliotis, V.L.; Chhabra, S.; Anderson, L.M.; Kyrtopoulos, S.A. Dosimetry of O6-methylguanine in rat DNA after low-dose, chronic exposure to N-nitrosodimethylamine (NDMA). Implications for the mechanism of NDMA hepatocarcinogenesis. Carcinogenesis 1995, 16, 2381–2387. [Google Scholar] [CrossRef]
- Montesano, R.; Brésil, H.; Planche-Martel, G.; Margison, G.P.; Pegg, A.E. Stability and capacity of dimethylnitrosamine-induced O6-methylguanine repair system in rat liver. Cancer Res. 1983, 43 Pt 1, 5808–5814. [Google Scholar]
- Hall, J.; Brésil, H.; Serres, M.; Martel-Planche, G.; Wild, C.P.; Montesano, R. Modulation of O6-methylguanine-DNA methyltransferase in rat and hamster liver after treatment with dimethylnitrosamine. Cancer Res. 1990, 50, 5426–5430. [Google Scholar] [PubMed]
- Lindamood, C., 3rd; Bedell, M.A.; Billings, K.C.; Dyroff, M.C.; Swenberg, J.A. Dose response for DNA alkylation, [3H]thymidine uptake into DNA, and O6-methylguanine-DNA methyltransferase activity in hepatocytes of rats and mice continuously exposed to dimethylnitrosamine. Cancer Res. 1984, 44, 196–200. [Google Scholar] [PubMed]
- Anderson, L.M.; Harrington, G.W.; Pylypiw, H.M., Jr.; Hagiwara, A.; Magee, P.N. Tissue levels and biological effects of N-nitrosodimethylamine in mice during chronic low or high dose exposure with or without ethanol. Drug Metab. Dispos. 1986, 14, 733–739. [Google Scholar] [PubMed]
- Chhabra, S.K.; Anderson, L.M.; Perella, C.; Desai, D.; Amin, S.; Kyrtopoulos, S.A.; Souliotis, V.L. Coexposure to ethanol with N-nitrosodimethylamine or 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone during lactation of rats: Marked increase in O6-methylguanine-DNA adducts in maternal mammary gland and in suckling lung and kidney. Toxicol. Appl. Pharmacol. 2000, 169, 191–200. [Google Scholar] [CrossRef]
- Belinsky, S.A.; Devereux, T.R.; Anderson, M.W. Role of DNA methylation in the activation of proto-oncogenes and the induction of pulmonary neoplasia by nitrosamines. Mutat. Res. 1990, 233, 105–116. [Google Scholar] [CrossRef]
- Nicoll, J.W.; Swann, P.F.; Pegg, A.E. Effect of dimethylnitrosamine on persistence of methylated guanines in rat liver and kidney DNA. Nature 1975, 254, 261–262. [Google Scholar] [CrossRef]
- Pegg, A.E.; Hui, G. Formation and subsequent removal of O6-methylguanine from deoxyribonucleic acid in rat liver and kidney after small doses of dimethylnitrosamine. Biochem. J. 1978, 173, 739–748. [Google Scholar] [CrossRef] [Green Version]
- Margison, G.P.; Kleihues, P. Chemical carcinogenesis in the nervous system. Preferential accumulation of O6-methylguanine in rat brain deoxyribonucleic acid during repetitive administration of N-methyl-N-nitrosourea. Biochem. J. 1975, 148, 521–525. [Google Scholar] [CrossRef] [Green Version]
- Loechler, E.L.; Green, C.L.; Essigmann, J.M. In vivo mutagenesis by O6-methylguanine built into a unique site in a viral genome. Proc. Natl. Acad. Sci. USA 1984, 81, 6271–6275. [Google Scholar] [CrossRef] [Green Version]
- Delaney, J.C.; Essigmann, J.M. Biological properties of single chemical-DNA adducts: A twenty year perspective. Chem. Res. Toxicol. 2008, 21, 232–252. [Google Scholar] [CrossRef] [Green Version]
- Pegg, A.E. Multifaceted roles of alkyltransferase and related proteins in DNA repair, DNA damage, resistance to chemotherapy, and research tools. Chem. Res. Toxicol. 2011, 24, 618–639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samson, L.; Han, S.; Marquis, J.C.; Rasmussen, L.J. Mammalian DNA repair methyltransferases shield O4MeT from nucleotide excision repair. Carcinogenesis 1997, 18, 919–924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, N.; Wang, J.; Wang, P.; Jiang, Y.; Wang, Y. In-vitro replication studies on O2-methylthymidine and O4-methylthymidine. Chem. Res. Toxicol. 2012, 25, 2523–2531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singer, B.; Sági, J.; Kuśmierek, J.T. Escherichia coli polymerase I can use O2-methyldeoxythymidine or O4-methyldeoxythymidine in place of deoxythymidine in primed poly(dA-dT).poly(dA-dT) synthesis. Proc. Natl. Acad. Sci. USA 1983, 80, 4884–4888. [Google Scholar] [CrossRef] [Green Version]
- Christov, P.P.; Yamanaka, K.; Choi, J.Y.; Takata, K.; Wood, R.D.; Guengerich, F.P.; Lloyd, R.S.; Rizzo, C.J. Replication of the 2,6-diamino-4-hydroxy-N5-(methyl)-formamidopyrimidine (MeFapy-dGuo) adduct by eukaryotic DNA polymerases. Chem. Res. Toxicol. 2012, 25, 1652–1661. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Wang, P.; Wang, Y. Cytotoxic and mutagenic properties of alkyl phosphotriester lesions in Escherichia coli cells. Nucleic Acids Res. 2018, 46, 4013–4021. [Google Scholar] [CrossRef] [Green Version]
- Harris, C.C.; Autrup, H.; Stoner, G.D.; Trump, B.F.; Hillman, E.; Schafer, P.W.; Jeffrey, A.M. Metabolism of benzo(a)pyrene, N-nitrosodimethylamine, and N-nitrosopyrrolidine and identification of the major carcinogen-DNA adducts formed in cultured human esophagus. Cancer Res. 1979, 39, 4401–4406. [Google Scholar]
- Herron, D.C.; Shank, R.C. Methylated purines in human liver DNA after probable dimethylnitrosamine poisoning. Cancer Res. 1980, 40, 3116–3117. [Google Scholar]
- Kyrtopoulos, S.A. DNA adducts in humans after exposure to methylating agents. Mutat. Res. 1998, 405, 135–143. [Google Scholar] [CrossRef]
- Phillips, D.H. Smoking-related DNA and protein adducts in human tissues. Carcinogenesis 2002, 23, 1979–2004. [Google Scholar] [CrossRef] [Green Version]
- Ma, B.; Stepanov, I.; Hecht, S.S. Recent studies on DNA adducts resulting from human exposure to tobacco smoke. Toxics 2019, 7, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foiles, P.G.; Miglietta, L.M.; Akerkar, S.A.; Everson, R.B.; Hecht, S.S. Detection of O6-methyldeoxyguanosine in human placental DNA. Cancer Res. 1988, 48, 4184–4188. [Google Scholar] [PubMed]
- Kang, H.I.; Konishi, C.; Eberle, G.; Rajewsky, M.F.; Kuroki, T.; Huh, N.H. Highly sensitive, specific detection of O6-methylguanine, O4-methylthymine, and O4-ethylthymine by the combination of high-performance liquid chromatography prefractionation, 32P postlabeling, and immunoprecipitation. Cancer Res. 1992, 52, 5307–5312. [Google Scholar] [PubMed]
- Kang, H.; Konishi, C.; Kuroki, T.; Huh, N. Detection of O6-methylguanine, O4-methylthymine and O4-ethylthymine in human liver and peripheral blood leukocyte DNA. Carcinogenesis 1995, 16, 1277–1280. [Google Scholar] [CrossRef]
- Umbenhauer, D.; Wild, C.P.; Montesano, R.; Saffhill, R.; Boyle, J.M.; Huh, N.; Kirstein, U.; Thomale, J.; Rajewsky, M.F.; Lu, S.H. O6-methyldeoxyguanosine in oesophageal DNA among individuals at high risk of oesophageal cancer. Int. J. Cancer 1985, 36, 661–665. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer. Some N-Nitroso Compounds. In IARC Monographs on the Evaluation of the Carcinogenic Risk of Chemicals to Humans; IARC: Lyon, France, 1978; Volume 17. [Google Scholar]
- Campillo, N.; Viñas, P.; Martínez-Castillo, N.; Hernández-Córdoba, M. Determination of volatile nitrosamines in meat products by microwave-assisted extraction and dispersive liquid-liquid microextraction coupled to gas chromatography-mass spectrometry. J. Chromatogr. A 2011, 1218, 1815–1821. [Google Scholar] [CrossRef]
- West, D.M.; Wu, Q.; Donovan, A.; Shi, H.; Ma, Y.; Jiang, H.; Wang, J. N-nitrosamine formation by monochloramine, free chlorine, and peracetic acid disinfection with presence of amine precursors in drinking water system. Chemosphere 2016, 153, 521–527. [Google Scholar] [CrossRef]
- Lim, H.H.; Oh, Y.S.; Shin, H.S. Determination of N-nitrosodimethylamine and N-nitrosomethylethylamine in drug substances and products of sartans, metformin and ranitidine by precipitation and solid phase extraction and gas chromatography-tandem mass spectrometry. J. Pharm. Biomed. Anal. 2020, 189, 113460. [Google Scholar] [CrossRef]
- Oldham, M.J.; DeSoi, D.J.; Rimmer, L.T.; Wagner, K.A.; Morton, M.J. Insights from analysis for harmful and potentially harmful constituents (HPHCs) in tobacco products. Regul. Toxicol. Pharmacol. 2014, 70, 138–148. [Google Scholar] [CrossRef]
- Lijinsky, W.; Reuber, M.D. Carcinogenicity in rats of nitrosomethylethylamines labeled with deuterium in several positions. Cancer Res. 1980, 40, 19–21. [Google Scholar]
- Lijinsky, W.; Saavedra, J.E.; Reuber, M.D.; Singer, S.S. Esophageal carcinogenesis in F344 rats by nitrosomethylethylamines substituted in the ethyl group. J. Natl. Cancer Inst. 1982, 68, 681–684. [Google Scholar] [PubMed]
- von Hofe, E.; Schmerold, I.; Nims, R.W.; Keefer, L.K.; Reist, E.J.; Kleihues, P. ß-deuteration of N-nitrosoethylmethylamine causes a shift in DNA methylation from rat liver to esophagus. Carcinogenesis 1991, 12, 545–549. [Google Scholar] [CrossRef] [PubMed]
- von Hofe, E.; Grahmann, F.; Keefer, L.K.; Lijinsky, W.; Nelson, V.; Kleihues, P. Methylation versus ethylation of DNA in target and nontarget tissues of Fischer 344 rats treated with N-nitrosomethylethylamine. Cancer Res. 1986, 46, 1038–1042. [Google Scholar] [PubMed]
- von Hofe, E.; Kleihues, P.; Keefer, L.K. Extent of DNA 2-hydroxyethylation by N-nitrosomethylethylamine and N-nitrosodiethylamine in vivo. Carcinogenesis 1986, 7, 1335–1337. [Google Scholar] [CrossRef] [Green Version]
- von Hofe, E.; Kleihues, P. Comparative studies on hepatic DNA alkylation in rats by N-nitrosomethylethylamine and N-nitrosodimethylamine plus N-nitrosodiethylamine. J. Cancer Res. Clin. Oncol. 1986, 112, 205–209. [Google Scholar] [CrossRef]
- National Toxicology Program. Report on Carcinogens, 12th ed.; U.S. Department of Health and Human Services, Public Health Service: Research Triangle Park, NC, USA, 2011.
- Sen, N.P.; Tessier, L.; Seaman, S.W. Determination of N-nitrosoproline and N-nitrososarcosine in malt and beer. J. Agric. Food Chem. 1983, 31, 1033–1036. [Google Scholar] [CrossRef]
- Wu, J.C.; Rickert, W.S.; Masters, A.; Joza, P. Determination of N-nitrososarcosine in tobacco and smokeless tobacco products using isotope dilution liquid chromatography tandem mass spectrometry. Anal. Methods 2012, 4, 3448–3452. [Google Scholar] [CrossRef]
- Werneth, M.; Pani, J.; Hofbauer, L.; Pummer, S.; Weber, M.T.; Pour, G.; Kahlig, H.; Mayer-Helm, B.; Stepan, H. Stereospecific response of E/Z-isomers of N-nitrososarcosine in LC-ESI-MS/MS. J. Chromatogr. Sci. 2021, 59, 813–822. [Google Scholar] [CrossRef]
- Derave, W.; Vanden Eede, E.; Hespel, P.; Carmella, S.G.; Hecht, S.S. Oral creatine supplementation in humans does not elevate urinary excretion of the carcinogen N-nitrososarcosine. Nutrition 2006, 22, 332–333. [Google Scholar] [CrossRef]
- Friedman, M.A. Reaction of sodium nitrite with dimethlglycine produces nitrososarcosine. Bull. Environ. Contam. Toxicol. 1975, 13, 226–232. [Google Scholar] [CrossRef]
- Friedman, M.A. Nitrosation of sarcosine: Chemical kinetics and gastric assay. Bull. Environ. Contam. Toxicol. 1972, 8, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.Y.; Wang, D.Y.; Tanaka, M.; Igarashi, H.; Kamo, T.; Shen, Q.; Sugimura, H.; Kino, I. Efficient and specific induction of esophageal tumors in rats by precursors of N-nitrososarcosine ethyl ester. Pathol. Int. 1995, 45, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Shubin, A.V.; Lesovaya, E.A.; Kirsanov, K.I.; Antoshina, E.E.; Trukhanova, L.S.; Gorkova, T.G.; Belitsky, G.A.; Yakubovskaya, M.G.; Demidyuk, I.V. Re-examination of the esophageal squamous cell carcinoma model in rats induced by N-nitrososarcosine ethyl ester precursors. Bull. Exp. Biol. Med. 2018, 164, 676–679. [Google Scholar] [CrossRef]
- Ohshima, H.; Bereziat, J.C.; Bartsch, H. Monitoring N-nitrosamino acids excreted in the urine and feces of rats as an index for endogenous nitrosation. Carcinogenesis 1982, 3, 115–120. [Google Scholar] [CrossRef]
- Cheng, G.; Wang, M.; Villalta, P.W.; Hecht, S.S. Detection of 7-(2′-carboxyethyl)guanine but not 7-carboxymethylguanine in human liver DNA. Chem. Res. Toxicol. 2010, 23, 1089–1096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, K.H.; Archer, M.C. Studies on the metabolic activation of ß-keto nitrosamines: Mechanisms of DNA methylation by N-(2-oxopropyl)-N-nitrosourea and N-nitroso-N-acetoxymethyl-N-2-oxopropylamine. Cell Biol. Int. 1984, 48, 169–179. [Google Scholar]
- Krüger, F.W. Metabolism of nitrosamines in vivo. I. Evidence for -oxydation of aliphatic di-n-alkylnitrosamines: The stimultaneous formation of 7-methylguanine besides 7-propyl- or 7-butylguanine after application of di-n-propyl- or di-n-butylnitrosamine. Z. Krebsforsch. Klin. Onkol. Cancer Res. Clin. Oncol. 1971, 76, 145–154. [Google Scholar] [CrossRef]
- Teiber, J.F.; Macé, K.; Hollenberg, P.F. Metabolism of the ß-oxidized intermediates of N-nitrosodi-n-propylamine: N-nitroso-ß-hydroxypropylpropylamine and N-nitroso-ß-oxopropylpropylamine. Carcinogenesis 2001, 22, 499–506. [Google Scholar] [CrossRef] [Green Version]
- Leung, K.H.; Archer, M.C. Mechanism of DNA methylation by N-nitroso(2-oxopropyl)propylamine. Carcinogenesis 1985, 6, 189–191. [Google Scholar] [CrossRef]
- Harrison, K.L.; Jukes, R.; Cooper, D.P.; Shuker, D.E. Detection of concomitant formation of O6-carboxymethyl- and O6-methyl-2′-deoxyguanosine in DNA exposed to nitrosated glycine derivatives using a combined immunoaffinity/HPLC method. Chem. Res. Toxicol. 1999, 12, 106–111. [Google Scholar] [CrossRef]
- Anderson, D.; Blowers, S.D. Limited cancer bioassay to test a potential food chemical. Lancet 1994, 344, 343–344. [Google Scholar] [CrossRef]
- Zurlo, J.; Curphey, T.J.; Hiley, R.; Longnecker, D.S. Identification of 7-carboxymethylguanine in DNA from pancreatic acinar cells exposed to azaserine. Cancer Res. 1982, 42, 1286–1288. [Google Scholar] [PubMed]
- Harrison, K.L.; Fairhurst, N.; Challis, B.C.; Shuker, D.E. Synthesis, characterization, and immunochemical detection of O6-(carboxymethyl)-2′-deoxyguanosine: A DNA adduct formed by nitrosated glycine derivatives. Chem. Res. Toxicol. 1997, 10, 652–659. [Google Scholar] [CrossRef] [PubMed]
- Shuker, D.E.; Margison, G.P. Nitrosated glycine derivatives as a potential source of O6-methylguanine in DNA. Cancer Res. 1997, 57, 366–369. [Google Scholar] [PubMed]
- Cupid, B.C.; Zeng, Z.; Singh, R.; Shuker, D.E. Detection of O6-carboxymethyl-2′-deoxyguanosine in DNA following reaction of nitric oxide with glycine and in human blood DNA using a quantitative immunoslot blot assay. Chem. Res. Toxicol. 2004, 17, 294–300. [Google Scholar] [CrossRef] [PubMed]
- Gottschalg, E.; Scott, G.B.; Burns, P.A.; Shuker, D.E. Potassium diazoacetate-induced p53 mutations in vitro in relation to formation of O6-carboxymethyl- and O6-methyl-2′-deoxyguanosine DNA adducts: Relevance for gastrointestinal cancer. Carcinogenesis 2007, 28, 356–362. [Google Scholar] [CrossRef] [Green Version]
- Jackson, P.E.; Hall, C.N.; Badawi, A.F.; O’Connor, P.J.; Cooper, D.P.; Povey, A.C. Frequency of Ki-ras mutations and DNA alkylation in colorectal tissue from individuals living in Manchester. Mol. Carcinog. 1996, 16, 12–19. [Google Scholar] [CrossRef]
- The EUROGAST Study Group. O6-Methylguanine in blood leucocyte DNA: An association with the geographic prevalence of gastric cancer and with low levels of serum pepsinogen A, a marker of severe chronic atrophic gastritis. Carcinogenesis 1994, 15, 1815–1820. [Google Scholar]
- Hall, C.N.; Badawi, A.F.; O’Connor, P.J.; Saffhill, R. The detection of alkylation damage in the DNA of human gastrointestinal tissues. Br. J. Cancer 1991, 64, 59–63. [Google Scholar] [CrossRef] [Green Version]
- Kyrtopoulos, S.A.; Vrotsou, B.; Golematis, B.; Bonatsos, M.; Lakiotis, G. O6-Methylguanine-DNA transmethylase activity in extracts of human gastric mucosa. Carcinogenesis 1984, 5, 943–947. [Google Scholar] [CrossRef]
- Povey, A.C.; Hall, C.N.; Badawi, A.F.; Cooper, D.P.; O’Connor, P.J. Elevated levels of the pro-carcinogenic adduct, O6-methylguanine, in normal DNA from the cancer prone regions of the large bowel. Gut 2000, 47, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.P.; Reed, P.I. N-nitroso compounds in fresh gastric juice and their relation to intragastric pH and nitrite employing an improved analytical method. Carcinogenesis 1993, 14, 2547–2551. [Google Scholar] [CrossRef] [PubMed]
- Bingham, S.A.; Pignatelli, B.; Pollock, J.R.; Ellul, A.; Malaveille, C.; Gross, G.; Runswick, S.; Cummings, J.H.; O’Neill, I.K. Does increased endogenous formation of N-nitroso compounds in the human colon explain the association between red meat and colon cancer? Carcinogenesis 1996, 17, 515–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Wang, Y. Chemical synthesis of oligodeoxyribonucleotides containing N3- and O4-carboxymethylthymidine and their formation in DNA. Nucleic Acids Res. 2009, 37, 336–345. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y. Synthesis and characterization of oligodeoxyribonucleotides containing a site-specifically incorporated N6-carboxymethyl-2′-deoxyadenosine or N4-carboxymethyl-2′-deoxycytidine. Nucleic Acids Res. 2010, 38, 6774–6784. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Wang, J.; Wang, P.; Wang, Y. Quantification of azaserine-induced carboxymethylated and methylated DNA lesions in cells by nanoflow liquid chromatography-nanoelectrospray ionization tandem mass spectrometry coupled with the stable isotope-dilution method. Anal. Chem. 2016, 88, 8036–8042. [Google Scholar] [CrossRef] [Green Version]
- Senthong, P.; Millington, C.L.; Wilkinson, O.J.; Marriott, A.S.; Watson, A.J.; Reamtong, O.; Eyers, C.E.; Williams, D.M.; Margison, G.P.; Povey, A.C. The nitrosated bile acid DNA lesion O6-carboxymethylguanine is a substrate for the human DNA repair protein O6-methylguanine-DNA methyltransferase. Nucleic Acids Res. 2013, 41, 3047–3055. [Google Scholar] [CrossRef]
- O’Driscoll, M.; Macpherson, P.; Xu, Y.Z.; Karran, P. The cytotoxicity of DNA carboxymethylation and methylation by the model carboxymethylating agent azaserine in human cells. Carcinogenesis 1999, 20, 1855–1862. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Leng, J.; Wang, Y. DNA replication studies of N-nitroso compound-induced O6-alkyl-2′-deoxyguanosine lesions in Escherichia coli. J. Biol. Chem. 2019, 294, 3899–3908. [Google Scholar] [CrossRef]
- Wu, J.; Wang, P.; Li, L.; Williams, N.L.; Ji, D.; Zahurancik, W.J.; You, C.; Wang, J.; Suo, Z.; Wang, Y. Replication studies of carboxymethylated DNA lesions in human cells. Nucleic Acids Res. 2017, 45, 7276–7284. [Google Scholar] [CrossRef] [Green Version]
- Swanson, A.L.; Wang, J.; Wang, Y. In vitro replication studies of carboxymethylated DNA lesions with Saccharomyces cerevisiae polymerase η. Biochemistry 2011, 50, 7666–7673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewin, M.H.; Bailey, N.; Bandaletova, T.; Bowman, R.; Cross, A.J.; Pollock, J.; Shuker, D.E.; Bingham, S.A. Red meat enhances the colonic formation of the DNA adduct O6-carboxymethyl guanine: Implications for colorectal cancer risk. Cancer Res. 2006, 66, 1859–1865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.S.; Yoo, J.S.; Ishizaki, H.; Hong, J.Y. Cytochrome P450IIE1: Roles in nitrosamine metabolism and mechanisms of regulation. Drug Metab. Rev. 1990, 22, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Camus, A.M.; Geneste, O.; Honkakoski, P.; Béréziat, J.C.; Henderson, C.J.; Wolf, C.R.; Bartsch, H.; Lang, M.A. High variability of nitrosamine metabolism among individuals: Role of cytochromes P450 2A6 and 2E1 in the dealkylation of N-nitrosodimethylamine and N-nitrosodiethylamine in mice and humans. Mol. Carcinog. 1993, 7, 268–275. [Google Scholar] [CrossRef]
- Verna, L.; Whysner, J.; Williams, G.M. N-nitrosodiethylamine mechanistic data and risk assessment: Bioactivation, DNA-adduct formation, mutagenicity, and tumor initiation. Pharmacol. Ther. 1996, 71, 57–81. [Google Scholar] [CrossRef]
- Singer, B. In vivo formation and persistence of modified nucleosides resulting from alkylating agents. Environ. Health Perspect. 1985, 62, 41–48. [Google Scholar] [CrossRef]
- Pegg, A.E. Alkylation and subsequent repair of DNA after exposure to dimethylnitrosamine and related carcinogens. Rev. Biochem. Toxicol. 1983, 38–133. [Google Scholar]
- Swenberg, J.A.; Dyroff, M.C.; Bedell, M.A.; Popp, J.A.; Huh, N.; Kirstein, U.; Rajewsky, M.F. O4-ethyldeoxythymidine, but not O6-ethyldeoxyguanosine, accumulates in hepatocyte DNA of rats exposed continuously to diethylnitrosamine. Proc. Natl. Acad. Sci. USA 1984, 81, 1692–1695. [Google Scholar] [CrossRef] [Green Version]
- Boucheron, J.A.; Richardson, F.C.; Morgan, P.H.; Swenberg, J.A. Molecular dosimetry of O4-ethyldeoxythymidine in rats continuously exposed to diethylnitrosamine. Cancer Res. 1987, 47, 1577–1581. [Google Scholar]
- Richardson, F.C.; Dyroff, M.C.; Boucheron, J.A.; Swenberg, J.A. Differential repair of O4-alkylthymidine following exposure to methylating and ethylating hepatocarcinogens. Carcinogenesis 1985, 6, 625–629. [Google Scholar] [CrossRef]
- Pegg, A.E. Properties of the O6-Alkylguanine-DNA Repair System of Mammalian Cells. In IARC Scientific Publication No. 57: N-Nitroso Compounds: Occurrence, Biological Effects and Relevance to Human Cancer; O’Neill, I., von Borstel, R., Miller, C., Long, J., Bartsch, H., Eds.; IARC: Lyon, France, 1984; pp. 575–580. [Google Scholar]
- Scherer, E.; Timmer, A.P.; Emmelot, P. Formation by diethylnitrosamine and persistence of O4-ethylthymidine in rat liver DNA in vivo. Cancer Lett. 1980, 10, 1–6. [Google Scholar] [CrossRef]
- Dolan, M.E.; Scicchitano, D.; Singer, B.; Pegg, A.E. Comparison of repair of methylated pyrimidines in poly(dT) by extracts from rat liver and Escherichia coli. Biochem. Biophys. Res. Commun. 1984, 123, 324–330. [Google Scholar] [CrossRef]
- Dragan, Y.P.; Hully, J.R.; Nakamura, J.; Mass, M.J.; Swenberg, J.A.; Pitot, H.C. Biochemical events during initiation of rat hepatocarcinogenesis. Carcinogenesis 1994, 15, 1451–1458. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Amato, N.J.; Zhai, Q.; Wang, Y. Cytotoxic and mutagenic properties of O4-alkylthymidine lesions in Escherichia coli cells. Nucleic Acids Res. 2015, 43, 10795–10803. [Google Scholar] [CrossRef] [Green Version]
- Graves, R.J.; Li, B.F.; Swann, P.F. Repair of O6-methylguanine, O6-ethylguanine, O6-isopropylguanine and O4-methylthymine in synthetic oligodeoxynucleotides by Escherichia coli ada gene O6-alkylguanine-DNA-alkyltransferase. Carcinogenesis 1989, 10, 661–666. [Google Scholar] [CrossRef]
- Wilkinson, M.C.; Potter, P.M.; Cawkwell, L.; Georgiadis, P.; Patel, D.; Swann, P.F.; Margison, G.P. Purification of the E. coli ogt gene product to homogeneity and its rate of action on O6-methylguanine, O6-ethylguanine and O4-methylthymine in dodecadeoxyribonucleotides. Nucleic Acids Res. 1989, 17, 8475–8484. [Google Scholar] [CrossRef]
- Saffhill, R. In vitro miscoding of alkylthymines with DNA and RNA polymerases. Cell Biol. Int. 1985, 53, 121–130. [Google Scholar] [CrossRef]
- Wu, J.; Li, L.; Wang, P.; You, C.; Williams, N.L.; Wang, Y. Translesion synthesis of O4-alkylthymidine lesions in human cells. Nucleic Acids Res. 2016, 44, 9256–9265. [Google Scholar]
- Singer, B.; Spengler, S.J.; Fraenkel-Conrat, H.; Kuśmierek, J.T. O4-Methyl, -ethyl, or -isopropyl substituents on thymidine in poly(dA-dT) all lead to transitions upon replication. Proc. Natl. Acad. Sci. USA 1986, 83, 28–32. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Wang, W.; Wu, J.; Shin, J.H.; Wang, P.; Unarta, I.C.; Chong, J.; Wang, Y.; Wang, D. Mechanism of DNA alkylation-induced transcriptional stalling, lesion bypass, and mutagenesis. Proc. Natl. Acad. Sci. USA 2017, 114, e7082–e7091. [Google Scholar] [CrossRef] [Green Version]
- Andersen, N.; Wang, P.; Wang, Y. Replication across regioisomeric ethylated thymidine lesions by purified DNA polymerases. Chem. Res. Toxicol. 2013, 26, 1730–1738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, N.L.; Wang, P.; Wang, Y. Replicative bypass of O2-alkylthymidine lesions in vitro. Chem. Res. Toxicol. 2016, 29, 1755–1761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, N.L.; Wang, P.; Wu, J.; Wang, Y. In vitro lesion bypass studies of O4-alkylthymidines with human DNA polymerase η. Chem. Res. Toxicol. 2016, 29, 669–675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhai, Q.; Wang, P.; Cai, Q.; Wang, Y. Syntheses and characterizations of the in vivo replicative bypass and mutagenic properties of the minor-groove O2-alkylthymidine lesions. Nucleic Acids Res. 2014, 42, 10529–10537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Wang, P.; Li, L.; You, C.; Wang, Y. Cytotoxic and mutagenic properties of minor-groove O2-alkylthymidine lesions in human cells. J. Biol. Chem. 2018, 293, 8638–8644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, R.; Kaur, B.; Farmer, P.B. Detection of DNA damage derived from a direct acting ethylating agent present in cigarette smoke by use of liquid chromatography-tandem mass spectrometry. Chem. Res. Toxicol. 2005, 18, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.J.; Wang, Y.C.; Lin, W.P. Analysis of ethylated thymidine adducts in human leukocyte DNA by stable isotope dilution nanoflow liquid chromatography-nanospray ionization tandem mass spectrometry. Anal. Chem. 2012, 84, 2521–2527. [Google Scholar] [CrossRef]
- Godschalk, R.; Nair, J.; Kliem, H.C.; Wiessler, M.; Bouvier, G.; Bartsch, H. Modified immunoenriched 32P-HPLC assay for the detection of O4-ethylthymidine in human biomonitoring studies. Chem. Res. Toxicol. 2002, 15, 433–437. [Google Scholar] [CrossRef]
- Godschalk, R.; Nair, J.; van Schooten, F.J.; Risch, A.; Drings, P.; Kayser, K.; Dienemann, H.; Bartsch, H. Comparison of multiple DNA adduct types in tumor adjacent human lung tissue: Effect of cigarette smoking. Carcinogenesis 2002, 23, 2081–2086. [Google Scholar] [CrossRef] [Green Version]
- Anna, L.; Kovács, K.; Gyorffy, E.; Schoket, B.; Nair, J. Smoking-related O4-ethylthymidine formation in human lung tissue and comparisons with bulky DNA adducts. Mutagenesis 2011, 26, 523–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.J.; Liu, Y.F. Simultaneous quantitative analysis of N3-ethyladenine and N7-ethylguanine in human leukocyte deoxyribonucleic acid by stable isotope dilution capillary liquid chromatography-nanospray ionization tandem mass spectrometry. J. Chromatogr. A 2013, 1271, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.J.; Lin, C.R. Noninvasive measurement of smoking-associated N3-ethyladenine and N7-ethylguanine in human salivary DNA by stable isotope dilution nanoflow liquid chromatography-nanospray ionization tandem mass spectrometry. Toxicol. Lett. 2014, 225, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Balbo, S.; Villalta, P.W.; Hecht, S.S. Quantitation of 7-ethylguanine in leukocyte DNA from smokers and nonsmokers by liquid chromatography-nanoelectrospray-high resolution tandem mass spectrometry. Chem. Res. Toxicol. 2011, 24, 1729–1734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prevost, V.; Shuker, D.E. Cigarette smoking and urinary 3-alkyladenine excretion in man. Chem. Res. Toxicol. 1996, 9, 439–444. [Google Scholar] [CrossRef]
- Kopplin, A.; Eberle-Adamkiewicz, G.; Glüsenkamp, K.H.; Nehls, P.; Kirstein, U. Urinary excretion of 3-methyladenine and 3-ethyladenine after controlled exposure to tobacco smoke. Carcinogenesis 1995, 16, 2637–2641. [Google Scholar] [CrossRef]
- Prevost, V.; Shuker, D.E.; Friesen, M.D.; Eberle, G.; Rajewsky, M.F.; Bartsch, H. Immunoaffinity purification and gas chromatography-mass spectrometric quantification of 3-alkyladenines in urine: Metabolism studies and basal excretion levels in man. Carcinogenesis 1993, 14, 199–204. [Google Scholar] [CrossRef]
- Carmella, S.G.; Chen, M.; Villalta, P.W.; Gurney, J.G.; Hatsukami, D.K.; Hecht, S.S. Ethylation and methylation of hemoglobin in smokers and non-smokers. Carcinogenesis 2002, 23, 1903–1910. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Wang, M.; Villalta, P.W.; Hecht, S.S. Liquid chromatography-electrospray ionization tandem mass spectrometry analysis of 7-ethylguanine in human liver DNA. Chem. Res. Toxicol. 2007, 20, 1498–1502. [Google Scholar] [CrossRef]
- Agency for Toxic Substances and Disease Registry. Toxicological Profile for N-Nitrosodi-n-Propylamine; Agency for Toxic Substances and Disease Registry: Atlanta, GA, USA, 2019.
- Shu, L.; Hollenberg, P.F. Identification of the cytochrome P450 isozymes involved in the metabolism of N-nitrosodipropyl-,N-nitrosodibutyl- and N-nitroso-n-butyl-n-propylamine. Carcinogenesis 1996, 17, 839–848. [Google Scholar] [CrossRef]
- Teiber, J.F.; Hollenberg, P.F. Identification of the human liver microsomal cytochrome P450s involved in the metabolism of N-nitrosodi-n-propylamine. Carcinogenesis 2000, 21, 1559–1566. [Google Scholar]
- Farrelly, J.G.; Stewart, M.L.; Lijinsky, W. The metabolism of nitrosodi-n-propylamine, nitrosodiallylamine and nitrosodiethanolamine. Carcinogenesis 1984, 5, 1015–1019. [Google Scholar] [CrossRef] [PubMed]
- Park, K.K.; Archer, M.C. Microsomal metabolism of N-nitrosodi-n-propylamine: Formation of products resulting from alpha- and beta-oxidation. Cell Biol. Int. 1978, 22, 83–90. [Google Scholar] [CrossRef]
- Park, K.K.; Wishnok, J.S.; Archer, M.C. Mechanism of alkylation by N-nitroso compounds: Detection of rearranged alcohol in the microsomal metabolism of N-nitrosodi-n-propylamine and base-catalyzed decomposition of N-n-propyl-N-nitrosourea. Cell Biol. Int. 1977, 18, 349–354. [Google Scholar] [CrossRef]
- Leung, K.H.; Archer, M.C. Urinary metabolites of N-nitrosodipropylamine, N-nitroso-2-hydroxypropylpropylamine and N-nitroso-2-oxopropylpropylamine in the rat. Carcinogenesis 1981, 2, 859–862. [Google Scholar] [CrossRef] [PubMed]
- Park, K.K.; Archer, M.C. Metabolism of N-nitroso-2-oxopropylpropylamine by rat liver: Formation of products resulting from both oxidation and reduction. Cancer Biochem. Biophys. 1978, 3, 37–40. [Google Scholar]
- Kokkinakis, D.M.; Hollenberg, P.F.; Scarpelli, D.G. Major urinary metabolites in hamsters and rats treated with N-nitroso(2-hydroxypropyl)(2-oxopropyl)amine. Cancer Res. 1985, 45, 3586–3592. [Google Scholar]
- Bauman, P.A.; Hotchkiss, J.H.; Parker, R.S. Metabolism of N-nitrosodi-n-propylamine and N-nitrosodiallylamine by isolated rat hepatocytes. Cancer Lett. 1985, 28, 229–236. [Google Scholar] [CrossRef]
- Appel, K.E.; Graf, H. Metabolic nitrite formation from N-nitrosamines: Evidence for a cytochrome P-450 dependent reaction. Carcinogenesis 1982, 3, 293–296. [Google Scholar] [CrossRef]
- Krüger, F.W. Metabolism of nitrosamines in vivo. II. On the methylation of nucleic acids by aliphatic di-n-alkyl-nitrosamines in vivo, caused by beta-oxydation: The increased formation of 7-methyl-guanine after application of beta-hydroxypropyl-propyl-nitrosamine compared to that after application of di-n-propyl-nitrosamine. Z. Krebsforsch. Klin. Onkol. Cancer Res. Clin. Oncol. 1973, 79, 90–97. [Google Scholar]
- Krüger, F.W.; Bertram, B. Metabolism of nitrosamines in vivo. 3. On the methylation of nucleic acids by aliphatic Di-n-alkyl-nitrosamines in vivo resulting from ß-oxidation: The formation of 7-methylguanine after application of 2-oxo-propyl-propyl-nitrosamine and methyl-propyl-nitro-samine. Z. Krebsforsch. Klin. Onkol. Cancer Res. Clin. Oncol. 1973, 80, 189–196. [Google Scholar]
- Shu, L.; Hollenberg, P.F. Alkylation of cellular macromolecules and target specificity of carcinogenic nitrosodialkylamines: Metabolic activation by cytochromes P450 2B1 and 2E1. Carcinogenesis 1997, 18, 801–810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kokkinakis, D.M. Alkylation of rodent tissue DNA induced by N-nitrosobis(2-hydroxypropyl)amine. Carcinogenesis 1992, 13, 759–765. [Google Scholar] [CrossRef] [PubMed]
- Kokkinakis, D.M. Differences between pancreatropic nitrosamine carcinogens and N-nitrosodimethylamine in methylating DNA in various tissues of hamsters and rats. Cell Biol. Int. 1991, 78, 167–181. [Google Scholar] [CrossRef]
- Kokkinakis, D.M. Differences in DNA-guanine alkylation between male Sprague-Dawley rats and Syrian hamsters following exposure to a single dose of pancreatic nitrosamine carcinogens. Chem. Res. Toxicol. 1990, 3, 150–156. [Google Scholar] [CrossRef]
- Fan, T.Y.; Morrison, J.; Rounbehler, D.P.; Ross, R.; Fine, D.H.; Miles, W.; Sen, N.P. N-Nitrosodiethanolamine in synthetic cutting fluids: A part-per-hundred impurity. Science 1977, 196, 70–71. [Google Scholar] [CrossRef]
- Fan, T.Y.; Goff, U.; Song, L.; Fine, D.H.; Arsenault, G.P.; Biemann, K. N-Nitrosodiethanolamine in cosmetics, lotions and shampoos. Food Cosmet. Toxicol. 1977, 15, 423–430. [Google Scholar] [CrossRef]
- Brunnemann, K.D.; Hoffmann, D. Assessment of the carcinogenic N-nitrosodiethanolamine in tobacco products and tobacco smoke. Carcinogenesis 1981, 2, 1123–1127. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer. Some Industrial Chemicals. In IARC Monographs on the Evaluation of Carcinogenic Risks to Humans; IARC: Lyon, France, 2000; Volume 77. [Google Scholar]
- Lethco, E.J.; Wallace, W.C.; Brouwer, E. The fate of N-nitrosodiethanolamine after oral and topical administration to rats. Food Chem. Toxicol. 1982, 20, 401–406. [Google Scholar] [CrossRef]
- Preussmann, R.; Wurtele, G.; Eisenbrand, G.; Spiegelhalder, B. Urinary excretion of N-nitrosodiethanolamine administered orally to rats. Cancer Lett. 1978, 4, 207–209. [Google Scholar] [CrossRef]
- Preussmann, R.; Habs, M.; Habs, H.; Schmähl, D. Carcinogenicity of N-nitrosodiethanolamine in rats at five different dose levels. Cancer Res. 1982, 42, 5167–5171. [Google Scholar]
- Hoffmann, D.; Rivenson, A.; Adams, J.D.; Juchatz, A.; Vinchkoski, N.; Hecht, S.S. A study of tobacco carcinogenesis. 24. Effects of route of administration and dose on the carcinogenicity of N-nitrosodiethanolamine in the Syrian Golden hamster. Cancer Res. 1983, 43, 2521–2524. [Google Scholar]
- Airoldi, L.; Bonfanti, M.; Benfenati, E.; Tavecchia, P.; Fanelli, R. Identification of an acidic metabolite of N-nitrosodiethanolamine isolated from rat urine. Biomed. Mass. Spectrom. 1983, 10, 334–337. [Google Scholar] [CrossRef] [PubMed]
- Farrelly, J.G.; Thomas, B.J.; Lijinsky, W. Metabolism and Cellular Interactions of N-Nitrosodiethanolamine. In IARC Scientific Publication No. 84: The Relevance of N-Nitroso Compounds to Human Cancer: Exposures and Mechanisms; Bartsch, H., O’Neill, I., Schulte-Hermann, R., Eds.; IARC: Lyon, France, 1987; pp. 87–90. [Google Scholar]
- Sterzel, W.; Eisenbrand, G. N-nitrosodiethanolamine is activated in the rat to an ultimate genotoxic metabolite by sulfotransferase. J. Cancer Res. Clin. Oncol. 1986, 111, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Michejda, C.J.; Andrews, A.W.; Koepke, S.R. Derivatives of side-chain hydroxylated nitrosamines direct acting mutagens in Salmonella typhimurium. Mutat. Res. 1979, 67, 301–308. [Google Scholar] [CrossRef]
- Loeppky, R.N.; Goelzer, P. Microsome-mediated oxidation of N-nitrosodiethanolamine (NDELA), a bident carcinogen. Chem. Res. Toxicol. 2002, 15, 457–469. [Google Scholar] [CrossRef]
- Hecht, S.S. N-Nitroso-2-hydroxymorpholine, a mutagenic metabolite of N-nitrosodiethanolamine. Carcinogenesis 1984, 5, 1745–1747. [Google Scholar] [CrossRef]
- Hecht, S.S.; Lijinsky, W.; Kovatch, R.M.; Chung, F.L.; Saavedra, J.E. Comparative tumorigenicity of N-nitroso-2-hydroxymorpholine, N-nitrosodiethanolamine and N-nitrosomorpholine in A/J mice and F344 rats. Carcinogenesis 1989, 10, 1475–1477. [Google Scholar] [CrossRef]
- Loeppky, R.N.; Ye, Q.; Goelzer, P.; Chen, Y. DNA adducts from N-nitrosodiethanolamine and related beta-oxidized nitrosamines in vivo: 32P-postlabeling methods for glyoxal- and O6-hydroxyethyldeoxyguanosine adducts. Chem. Res. Toxicol. 2002, 15, 470–482. [Google Scholar] [CrossRef]
- Loeppky, R.N.; Cui, W.; Goelzer, P.; Park, M.; Ye, Q. Glyoxal-guanine DNA Adducts: Detection, Stability and Formation in vivo from Nitrosamines. In IARC Scientific Publication No. 150: Exocyclic DNA Adducts in Mutagenesis and Carcinogenesis; Singer, B., Bartsch, H., Eds.; IARC: Lyon, France, 1999; pp. 155–168. [Google Scholar]
- Loeppky, R.N.; Fuchs, A.; Janzowski, C.; Humberd, C.; Goelzer, P.; Schneider, H.; Eisenbrand, G. Probing the mechanism of the carcinogenic activation of N-nitrosodiethanolamine with deuterium isotope effects: In Vivo induction of DNA single-strand breaks and related in vitro assays. Chem. Res. Toxicol. 1998, 11, 1556–1566. [Google Scholar] [CrossRef]
- Loeppky, R.N. The mechanism of bioactivation of N-nitrosodiethanolamine. Drug Metab. Rev. 1999, 31, 175–193. [Google Scholar] [CrossRef]
- Loeppky, R.N.; Sukhtankar, S.; Gu, F.; Park, M. The carcinogenic significance of reactive intermediates derived from 3-acetoxy- and 5-acetoxy-2-hydroxy-N-nitrosomorpholine. Chem. Res. Toxicol. 2005, 18, 1955–1966. [Google Scholar] [CrossRef] [PubMed]
- Dennehy, M.K.; Loeppky, R.N. Mass spectrometric methodology for the determination of glyoxaldeoxyguanosine and O6-hydroxyethyldeoxyguanosine DNA adducts produced by nitrosamine bident carcinogens. Chem. Res. Toxicol. 2005, 18, 556–565. [Google Scholar] [CrossRef]
- Giménez-Campillo, C.; Pastor-Belda, M.; Campillo, N.; Hernández-Córdoba, M.; Viñas, P. Development of a new methodology for the determination of N-nitrosamines impurities in ranitidine pharmaceuticals using microextraction and gas chromatography-mass spectrometry. Talanta 2021, 223 Pt 2, 121659. [Google Scholar] [CrossRef] [PubMed]
- Imaida, K.; Wang, C.Y. Effect of sodium phenobarbital and sodium saccharin in AIN-76A diet on carcinogenesis initiated with N-[4-(5-nitro-2-furyl)-2-thiazolyl]formamide and N,N-dibutylnitrosamine in male F344 rats. Cancer Res. 1986, 46 Pt 1, 6160–6164. [Google Scholar] [PubMed]
- Fukushima, S.; Sakata, T.; Tagawa, Y.; Shibata, M.A.; Hirose, M.; Ito, N. Different modifying response of butylated hydroxyanisole, butylated hydroxytoluene, and other antioxidants in N,N-dibutylnitrosamine esophagus and forestomach carcinogenesis of rats. Cancer Res. 1987, 47, 2113–2116. [Google Scholar]
- Imaida, K.; Fukushima, S.; Inoue, K.; Masui, T.; Hirose, M.; Ito, N. Modifying effects of concomitant treatment with butylated hydroxyanisole or butylated hydroxytoluene on N,N-dibutylnitrosamine-induced liver, forestomach and urinary bladder carcinogenesis in F344 male rats. Cancer Lett. 1988, 43, 167–172. [Google Scholar] [CrossRef]
- Hirose, M.; Uwagawa, S.; Ozaki, K.; Takaba, K.; Ito, N. Effects of butylated hydroxyanisole pretreatment on low dose N-methyl-N′-nitro-N-nitrosoguanidine- or N,N-dibutylnitrosamine-induced rat forestomach or esophageal carcinogenesis. Carcinogenesis 1991, 12, 1773–1776. [Google Scholar] [CrossRef] [PubMed]
- Shibata, M.A.; Fukushima, S.; Takahashi, S.; Hasegawa, R.; Ito, N. Enhancing effects of sodium phenobarbital and N,N-dibutylnitrosamine on tumor development in a rat wide-spectrum organ carcinogenesis model. Carcinogenesis 1990, 11, 1027–1031. [Google Scholar] [CrossRef]
- Lijinsky, W.; Reuber, M.D. Carcinogenesis in Fischer rats by nitrosodipropylamine, nitrosodibutylamine and nitrosobis(2-oxopropyl)amine given by gavage. Cancer Lett. 1983, 19, 207–213. [Google Scholar] [CrossRef]
- Tsuda, H.; Mera, Y.; Seki, K.; Aoki, T.; Fukushima, S.; Ito, N. Induction of tumors in the liver, urinary bladder, esophagus and forestomach by short-term treatment with different doses of N,N′-dibutylnitrosamine in rats. Jpn. J. Cancer Res. 1987, 78, 227–234. [Google Scholar]
- Nishikawa, T.; Salim, E.I.; Morimura, K.; Kaneko, M.; Ogawa, M.; Kinoshita, A.; Osugi, H.; Kinoshita, H.; Fukushima, S. High susceptibility of p53 knockout mice to esophageal and urinary bladder carcinogenesis induced by N,N-dibutylnitrosamine. Cancer Lett. 2003, 194, 45–54. [Google Scholar] [CrossRef]
- Suzuki, E.; Mochizuki, M.; Wakabayashi, Y.; Okada, M. In vitro metabolic activation of N,N-dibutylnitrosamine in mutagenesis. Gan 1983, 74, 51–59. [Google Scholar]
- Suzuki, E.; Mochizuki, M.; Shibuya, K.; Okada, M. N,N-dibutylnitrosamine metabolism by rat liver S9 fraction and oxidation by chemical model systems. Gan 1983, 74, 41–50. [Google Scholar] [PubMed]
- Suzuki, E.; Okada, M. Metabolic fate of N,N-dibutylnitrosamine in the rat. Gan 1980, 71, 863–870. [Google Scholar] [PubMed]
- Vasconcelos-Nóbrega, C.; Colaço, A.; Lopes, C.; Oliveira, P.A. Review: BBN as an urothelial carcinogen. In Vivo 2012, 26, 727–739. [Google Scholar] [PubMed]
- Suzuki, E.; Okada, M. Metabolic fate of N-butyl-N-(4-hydroxybutyl)nitrosamine in the rat. Gan 1980, 71, 856–862. [Google Scholar] [PubMed]
- Bonfanti, M.; Magagnotti, C.; Bonati, M.; Fanelli, R.; Airoldi, L. Pharmacokinetic profile and metabolism of N-nitrosobutyl-(4-hydroxybutyl)amine in rats. Cancer Res. 1988, 48, 3666–3669. [Google Scholar] [PubMed]
- Suzuki, E.; Aoki, J.; Okada, M. Metabolic fate of N-butyl-N-(4-hydroxybutyl) nitrosamine and N,N-dibutylnitrosamine in the guinea pig, with reference to their carcinogenic effects on the urinary bladder. Gan 1981, 72, 547–551. [Google Scholar]
- Suzuki, E.; Osabe, M.; Okada, M.; Ishizaka, T.; Suzuki, T. Urinary metabolites of N-nitrosodibutylamine and N-nitrodibutylamine in the rat: Identification of N-acetyl-S-alkyl-L-cysteines. Jpn. J. Cancer Res. 1987, 78, 382–385. [Google Scholar]
- Bonfanti, M.; Magagnotti, C.; Galli, A.; Bagnati, R.; Moret, M.; Gariboldi, P.; Fanelli, R.; Airoldi, L. Determination of O6-butylguanine in DNA by immunoaffinity extraction/gas chromatography-mass spectrometry. Cancer Res. 1990, 50, 6870–6875. [Google Scholar]
- Airoldi, L.; Magagnotti, C.; Bonfanti, M.; Chiappetta, L.; Lolli, M.; Medana, C.; De Gregorio, G.; Fanelli, R. Detection of O6-butyl- and O6-(4-hydroxybutyl)guanine in urothelial and hepatic DNA of rats given the bladder carcinogen N-nitrosobutyl(4-hydroxybutyl)amine. Carcinogenesis 1994, 15, 2297–2301. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, D.; Brunnemann, K.D.; Adams, J.D.; Hecht, S.S. Formation and Analysis of N-Nitrosamines in Tobacco Products and their Endogenous Formation in Consumers. In IARC Scientific Publication No. 57: N-Nitroso Compounds: Occurrence, Biological Effects and Relevance to Human Cancer; O’Neill, I., von Borstel, R., Miller, C., Long, J., Bartsch, H., Eds.; IARC: Lyon, France, 1984; pp. 743–762. [Google Scholar]
- Tricker, A.R.; Pfundstein, B.; Kälble, T.; Preussmann, R. Secondary amine precursors to nitrosamines in human saliva, gastric juice, blood, urine and faeces. Carcinogenesis 1992, 13, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Lijinsky, W.; Reuber, M.D. Carcinogenic effect of nitrosopyrrolidine, nitrosopiperidine and nitrosohexamethyleneimine in Fischer rats. Cancer Lett. 1981, 12, 99–103. [Google Scholar] [CrossRef]
- Chung, F.L.; Tanaka, T.; Hecht, S.S. Induction of liver tumors in F344 rats by crotonaldehyde. Cancer Res. 1986, 46, 1285–1289. [Google Scholar] [PubMed]
- Gray, R.; Peto, R.; Brantom, P.; Grasso, P. Chronic nitrosamine ingestion in 1040 rodents: The effect of the choice of nitrosamine, the species studied, and the age of starting exposure. Cancer Res. 1991, 51 Pt 2, 6470–6491. [Google Scholar] [PubMed]
- Hecht, S.S.; Abbaspour, A.; Hoffman, D. A study of tobacco carcinogenesis XLII. Bioassay in mice of some structural analogues of tobacco-specific nitrosamines. Cancer Lett. 1988, 42, 141–145. [Google Scholar] [CrossRef]
- McCoy, G.D.; Hecht, S.S.; Katayama, S.; Wynder, E.L. Differential effect of chronic ethanol consumption on the carcinogenicity of N-nitrosopyrrolidine and N′-nitrosonornicotine in male Syrian Golden hamsters. Cancer Res. 1981, 41, 2849–2854. [Google Scholar]
- McCoy, G.D.; Hecht, S.S.; Furuya, K. The effect of chronic ethanol consumption on the tumorigenicity of N-nitrosopyrrolidine in male Syrian golden hamsters. Cancer Lett. 1986, 33, 151–159. [Google Scholar] [CrossRef]
- Hecht, S.S.; McCoy, G.D.; Chen, C.-H.B.; Hoffmann, D. The Metabolism of Cyclic Nitrosamines. In N-Nitroso Compounds; Scanlan, R.A., Tannenbaum, S.R., Eds.; American Chemical Society: Washington, DC, USA, 1981; Volume 174, pp. 49–75. [Google Scholar]
- Snyder, C.M.; Farrelly, J.G.; Lijinsky, W. Metabolism of three cyclic nitrosamines in Sprague-Dawley rats. Cancer Res. 1977, 37, 3530–3532. [Google Scholar]
- Wong, H.L.; Murphy, S.E.; Hecht, S.S. Cytochrome P450 2A-catalyzed metabolic activation of structurally similar carcinogenic nitrosamines: N′-nitrosonornicotine enantiomers, N-nitrosopiperidine, and N-nitrosopyrrolidine. Chem. Res. Toxicol. 2005, 18, 61–69. [Google Scholar] [CrossRef]
- Hecht, S.S.; Chen, C.B.; Hoffmann, D. Evidence for metabolic α hydroxylation of N-nitrosopyrrolidine. Cancer Res. 1978, 38, 215–218. [Google Scholar] [PubMed]
- Chen, C.B.; McCoy, G.D.; Hecht, S.S.; Hoffmann, D.; Wynder, E.L. High pressure liquid chromatographic assay for alpha hydroxylation of N-nitrosopyrrolidine by isolated rat liver microsomes. Cancer Res. 1978, 38 Pt 1, 3812–3816. [Google Scholar] [PubMed]
- Hecker, L.I.; Farrelly, J.G.; Smith, J.H.; Saavedra, J.E.; Lyon, P.A. Metabolism of the liver carcinogen N-nitrosopyrrolidine by rat liver microsomes. Cancer Res. 1979, 39 Pt 1, 2679–2686. [Google Scholar] [PubMed]
- Wang, M.Y.; Chung, F.L.; Hecht, S.S. Identification of crotonaldehyde as a hepatic microsomal metabolite formed by α-hydroxylation of the carcinogen N-nitrosopyrrolidine. Chem. Res. Toxicol. 1988, 1, 28–31. [Google Scholar] [CrossRef]
- Krüger, F.W.; Bertram, B. Metabolism of nitrosamines in vivo IV. Isolation of 3-hydroxy-1-nitrosopyrrolidine from rat urine after application of 1-nitrosopyrrolidine. Z. Krebsforsch. Klin. Onkol. Cancer Res. Clin. Oncol. 1975, 83, 255–260. [Google Scholar] [CrossRef]
- Cottrell, R.C.; Walters, D.G.; Young, P.J.; Phillips, J.C.; Lake, B.G.; Gangolli, S.D. Studies of the urinary metabolites of N-nitrosopyrrolidine in the rat. Toxicol. Appl. Pharmacol. 1980, 54, 368–376. [Google Scholar] [CrossRef]
- Wang, M.; McIntee, E.J.; Cheng, G.; Shi, Y.; Villalta, P.W.; Hecht, S.S. Identification of paraldol-deoxyguanosine adducts in DNA reacted with crotonaldehyde. Chem. Res. Toxicol. 2000, 13, 1065–1074. [Google Scholar] [CrossRef]
- Wang, M.; McIntee, E.J.; Cheng, G.; Shi, Y.; Villalta, P.W.; Hecht, S.S. A Schiff base is a major DNA adduct of crotonaldehyde. Chem. Res. Toxicol. 2001, 14, 423–430. [Google Scholar] [CrossRef]
- Wang, M.; Upadhyaya, P.; Dinh, T.T.; Bonilla, L.E.; Hecht, S.S. Lactols in hydrolysates of DNA treated with α-acetoxy-N-nitrosopyrrolidine or crotonaldehyde. Chem. Res. Toxicol. 1998, 11, 1567–1573. [Google Scholar] [CrossRef]
- Autrup, H.; Stoner, G.D. Metabolism of N-nitrosamines by cultured human and rat esophagus. Cancer Res. 1982, 42, 1307–1311. [Google Scholar]
- Autrup, H.; Harris, C.C.; Trump, B.F. Metabolism of acyclic and cyclic N-nitrosamines by cultured human colon. Proc. Soc. Exp. Biol. Med. 1978, 159, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Autrup, H.; Grafstrom, R.C.; Christensen, B.; Kieler, J. Metabolism of chemical carcinogens by cultured human and rat bladder epithelial cells. Carcinogenesis 1981, 2, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Hecht, S.S.; Chen, C.H.; McCoy, G.D.; Hoffmann, D.; Domellof, L. α-hydroxylation of N-nitrosopyrrolidine and N′-nitrosonornicotine by human liver microsomes. Cancer Lett. 1979, 8, 35–41. [Google Scholar] [CrossRef]
- Hecker, L.I. In vitro metabolism of N-nitrosopyrrolidine by rat lung subcellular fractions: Alpha-hydroxylation in a non-target tissue. Cell Biol. Int. 1980, 30, 57–65. [Google Scholar] [CrossRef]
- McCoy, G.D.; Chen, C.H.; Hecht, S.S.; McCoy, E.C. Enhanced metabolism and mutagenesis of nitrosopyrrolidine in liver fractions isolated from chronic ethanol-consuming hamsters. Cancer Res. 1979, 39, 793–796. [Google Scholar] [PubMed]
- McCoy, G.D.; Hecht, S.S.; McCoy, E.C. Comparison of microsomal inducer pretreatment on the in vitro alpha-hydroxylation and mutagenicity of N-Nitrosopyrrolidine in rat and hamster liver. Environ. Mutagen. 1982, 4, 221–230. [Google Scholar] [CrossRef]
- Hunt, E.J.; Shank, R.C. Evidence for DNA adducts in rat liver after adminstration of N-nitrosopyrrolidine. Biochem. Biophys. Res. Commun. 1982, 104, 1343–1348. [Google Scholar] [CrossRef]
- Chung, F.L.; Wang, M.Y.; Hecht, S.S. Detection of exocyclic guanine adducts in hydrolysates of hepatic DNA of rats treated with N-nitrosopyrrolidine and in calf thymus DNA reacted with alpha-acetoxy-N-nitrosopyrrolidine. Cancer Res. 1989, 49, 2034–2041. [Google Scholar]
- Wang, M.; Chung, F.L.; Hecht, S.S. Formation of acyclic and cyclic guanine adducts in DNA reacted with alpha-acetoxy-N-nitrosopyrrolidine. Chem. Res. Toxicol. 1989, 2, 423–428. [Google Scholar] [CrossRef]
- Wang, M.; Nishikawa, A.; Chung, F.L. Differential effects of thiols on DNA modifications via alkylation and Michael addition by α-acetoxy-N-nitrosopyrrolidine. Chem. Res. Toxicol. 1992, 5, 528–531. [Google Scholar] [CrossRef]
- Loureiro, A.P.; Zhang, W.; Kassie, F.; Zhang, S.; Villalta, P.W.; Wang, M.; Hecht, S.S. Mass spectrometric analysis of a cyclic 7,8-butanoguanine adduct of N-nitrosopyrrolidine: Comparison to other N-nitrosopyrrolidine adducts in rat hepatic DNA. Chem. Res. Toxicol. 2009, 22, 1728–1735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunt, E.J.; Shank, R.C. Formation and persistence of a DNA adduct in rodents treated with N-nitrosopyrrolidine. Carcinogenesis 1991, 12, 571–575. [Google Scholar] [CrossRef]
- Wang, M.; Hecht, S.S. A cyclic N7,C-8 guanine adduct of N-nitrosopyrrolidine (NPYR): Formation in nucleic acids and excretion in the urine of NPYR-treated rats. Chem. Res. Toxicol. 1997, 10, 772–778. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Chung, F.L.; Hecht, S.S. Formation of 7-(4-oxobutyl)guanine in hepatic DNA of rats treated with N-nitrosopyrrolidine. Carcinogenesis 1992, 13, 1909–1911. [Google Scholar] [CrossRef] [PubMed]
- Chung, F.L.; Young, R.; Hecht, S.S. Detection of cyclic 1,N2-propanodeoxyguanosine adducts in DNA of rats treated with N-nitrosopyrrolidine and mice treated with crotonaldehyde. Carcinogenesis 1989, 10, 1291–1297. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Lao, Y.; Cheng, G.; Shi, Y.; Villalta, P.W.; Nishikawa, A.; Hecht, S.S. Analysis of adducts in hepatic DNA of rats treated with N-nitrosopyrrolidine. Chem. Res. Toxicol. 2007, 20, 634–640. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Young-Sciame, R.; Chung, F.L.; Hecht, S.S. Formation of N2-tetrahydrofuranyl and N2-tetrahydropyranyl adducts in the reactions of α-acetoxy-N-nitrosopyrrolidine and α-acetoxy-N-nitrosopiperidine with DNA. Chem. Res. Toxicol. 1995, 8, 617–624. [Google Scholar] [CrossRef]
- Young-Sciame, R.; Wang, M.; Chung, F.L.; Hecht, S.S. Reactions of α-acetoxy-N-nitrosopyrrolidine and α-acetoxy-N-nitrosopiperidine with deoxyguanosine: Formation of N2-tetrahydrofuranyl and N2-tetrahydropyranyl adducts. Chem. Res. Toxicol. 1995, 8, 607–616. [Google Scholar] [CrossRef]
- Wang, M.; Lao, Y.; Cheng, G.; Shi, Y.; Villalta, P.W.; Hecht, S.S. Identification of adducts formed in the reaction of α-acetoxy-N-nitrosopyrrolidine with deoxyribonucleosides and DNA. Chem. Res. Toxicol. 2007, 20, 625–633. [Google Scholar] [CrossRef] [Green Version]
- Hermida, S.A.; Possari, E.P.; Souza, D.B.; de Arruda Campos, I.P.; Gomes, O.F.; Di Mascio, P.; Medeiros, M.H.; Loureiro, A.P. 2′-Deoxyguanosine, 2′-deoxycytidine, and 2′-deoxyadenosine adducts resulting from the reaction of tetrahydrofuran with DNA bases. Chem. Res. Toxicol. 2006, 19, 927–936. [Google Scholar] [CrossRef]
- Chung, F.L.; Hecht, S.S. Formation of cyclic 1,N2-adducts by reaction of deoxyguanosine with alpha-acetoxy-N-nitrosopyrrolidine, 4-(carbethoxynitrosamino)butanal, or crotonaldehyde. Cancer Res. 1983, 43, 1230–1235. [Google Scholar] [PubMed]
- Chung, F.L.; Roy, K.R.; Hecht, S.S. A study of chemical carcinogenesis. 104. A study of reactions of α,ß-unsaturated carbonyl-compounds with deoxyguanosine. J. Org. Chem. 1988, 53, 14–17. [Google Scholar] [CrossRef]
- Wang, M.; McIntee, E.J.; Shi, Y.; Cheng, G.; Upadhyaya, P.; Villalta, P.W.; Hecht, S.S. Reactions of α-acetoxy-N-nitrosopyrrolidine with deoxyguanosine and DNA. Chem. Res. Toxicol. 2001, 14, 1435–1445. [Google Scholar] [CrossRef] [PubMed]
- Chahoua, L.; Cai, H.; Fishbein, J.C. Cyclic α-acetoxynitrosamines: Mechanisms of decomposition and stability of α-hydroxynitrosamine and nitrosiminium ion reactive intermediates. J. Am. Chem. Soc. 1999, 121, 5161–5169. [Google Scholar] [CrossRef]
- Elespuru, R.K.; Lijinsky, W. Mutagenicity of cyclic nitrosamines in Escherichia coli following activation with rat liver microsomes. Cancer Res. 1976, 36 Pt 1, 4099–4101. [Google Scholar]
- Gomez, R.F.; Johnston, M.; Sinskey, A.J. Activation of nitrosomorpholine and nitrosopyrrolidine to bacterial mutagens. Mutat. Res. 1974, 24, 5–7. [Google Scholar] [CrossRef]
- Wang, H.T.; Zhang, S.; Hu, Y.; Tang, M.S. Mutagenicity and sequence specificity of acrolein-DNA adducts. Chem. Res. Toxicol. 2009, 22, 511–517. [Google Scholar] [CrossRef] [Green Version]
- Tang, M.S.; Wang, H.T.; Hu, Y.; Chen, W.S.; Akao, M.; Feng, Z.; Hu, W. Acrolein induced DNA damage, mutagenicity and effect on DNA repair. Mol. Nutr. Food. Res. 2011, 55, 1291–1300. [Google Scholar] [CrossRef] [Green Version]
- Minko, I.G.; Kozekov, I.D.; Harris, T.M.; Rizzo, C.J.; Lloyd, R.S.; Stone, M.P. Chemistry and biology of DNA containing 1,N2-deoxyguanosine adducts of the α,ß-unsaturated aldehydes acrolein, crotonaldehyde, and 4-hydroxynonenal. Chem. Res. Toxicol. 2009, 22, 759–778. [Google Scholar] [CrossRef] [Green Version]
- Nair, U.; Bartsch, H.; Nair, J. Lipid peroxidation-induced DNA damage in cancer-prone inflammatory diseases: A review of published adduct types and levels in humans. Free Radic. Biol. Med. 2007, 43, 1109–1120. [Google Scholar] [CrossRef]
- Chung, F.L.; Nath, R.G.; Nagao, M.; Nishikawa, A.; Zhou, G.D.; Randerath, K. Endogenous formation and significance of 1,N2-propanodeoxyguanosine adducts. Mutat. Res. 1999, 424, 71–81. [Google Scholar] [CrossRef]
- Zhang, S.; Villalta, P.W.; Wang, M.; Hecht, S.S. Analysis of crotonaldehyde- and acetaldehyde-derived 1,N2-propanodeoxyguanosine adducts in DNA from human tissues using liquid chromatography electrospray ionization tandem mass spectrometry. Chem. Res. Toxicol. 2006, 19, 1386–1392. [Google Scholar] [CrossRef] [Green Version]
- Chung, F.L.; Chen, H.J.; Nath, R.G. Lipid peroxidation as a potential endogenous source for the formation of exocyclic DNA adducts. Carcinogenesis 1996, 17, 2105–2111. [Google Scholar] [CrossRef] [PubMed]
- De Mey, E.; De Klerck, K.; De Maere, H.; Dewulf, L.; Derdelinckx, G.; Peeters, M.C.; Fraeye, I.; Vander Heyden, Y.; Paelinck, H. The occurrence of N-nitrosamines, residual nitrite and biogenic amines in commercial dry fermented sausages and evaluation of their occasional relation. Meat Sci. 2014, 96 Pt A, 821–828. [Google Scholar] [CrossRef]
- De Mey, E.; De Maere, H.; Goemaere, O.; Steen, L.; Peeters, M.C.; Derdelinckx, G.; Paelinck, H.; Fraeye, I. Evaluation of N-nitrosopiperidine formation from biogenic amines during the production of dry fermented sausages. Food Bioproc. Technol. 2013, 7, 1269–1280. [Google Scholar] [CrossRef]
- Gilbert, P.J.; Rollmann, B.; Rondelet, J.; Mercier, M.; Poncelet, F. Mutagenicity and alpha-hydroxylation of N-nitrosopyrrolidine and N-nitrosopiperidine: A possible correlation. Toxicology 1981, 22, 345–352. [Google Scholar] [CrossRef]
- Ayrton, A.D.; Smith, J.N.; Ioannides, C. Bioactivation of N-nitrosopiperidine to mutagens: Role of hepatic cytochrome P-450 proteins and contribution of cytosolic fraction. Carcinogenesis 1987, 8, 1691–1695. [Google Scholar] [CrossRef]
- Lai, D.Y.; Arcos, J.C.; Argus, M.F. Factors influencing the microsome- and mitochondria-catalyzed in vitro binding of diethylnitrosamine and N-nitrosopiperidine to deoxyribonucleic acid. Biochem. Pharmacol. 1979, 28, 3545–3550. [Google Scholar] [CrossRef]
- Wong, H.L.; Murphy, S.E.; Hecht, S.S. Preferential metabolic activation of N-nitrosopiperidine as compared to its structural homologue N-nitrosopyrrolidine by rat nasal mucosal microsomes. Chem. Res. Toxicol. 2003, 16, 1298–1305. [Google Scholar] [CrossRef]
- Wong, H.L.; Murphy, S.E.; Wang, M.; Hecht, S.S. Comparative metabolism of N-nitrosopiperidine and N-nitrosopyrrolidine by rat liver and esophageal microsomes and cytochrome P450 2A3. Carcinogenesis 2003, 24, 291–300. [Google Scholar] [CrossRef] [Green Version]
- Arimoto, S.; Shimada, H.; Ukawa, S.; Mochizuki, M.; Hayatsu, H. Formation of directly mutagenic alpha-hydroxy-N-nitrosopiperidine phosphate ester by near-ultraviolet irradiation of N-nitrosopiperidine in phosphate buffer. Biochem. Biophys. Res. Commun. 1989, 162, 1140–1146. [Google Scholar] [CrossRef] [Green Version]
- Hecht, S.S.; Young-Sciame, R.; Chung, F.L. Reaction of α-acetoxy-N-nitrosopiperidine with deoxyguanosine: Oxygen-dependent formation of 4-oxo-2-pentenal and a 1,N2-ethenodeoxyguanosine adduct. Chem. Res. Toxicol. 1992, 5, 706–712. [Google Scholar] [CrossRef] [PubMed]
- Leung, K.H.; Park, K.K.; Archer, M.C. Alpha-hydroxylation in the metabolism of N-nitrosopiperidine by rat liver microsomes: Formation of 5-hydroxypentanal. Res. Commun. Chem. Pathol. Pharmacol. 1978, 19, 201–211. [Google Scholar] [PubMed]
- Nakamura, M.; Horiguchi, Y.; Kawabata, T. Effect of Ascorbic Acid and Some Reducing Agents on N-Nitrosopiperidine Metabolism by Liver Microsomes. In IARC Scientific Publication No. 57: N-Nitroso Compounds: Occurrence, Biological Effects and Relevance to Human Cancer; O’Neill, I., von Borstel, R., Miller, C., Long, J., Bartsch, H., Eds.; IARC: Lyon, France, 1984; pp. 547–552. [Google Scholar]
- Ravindranath, V.; Burka, L.T.; Boyd, M.R. Reactive metabolites from the bioactivation of toxic methylfurans. Science 1984, 224, 884–886. [Google Scholar] [CrossRef] [PubMed]
- Rayman, M.P.; Challis, B.C. Oxidation of N-nitrosopiperidine in the Udenfriend model system and its metabolism by rat-liver microsomes. Biochem. Pharmacol. 1975, 24, 621–626. [Google Scholar] [CrossRef]
- Challis, B.C.; Rayman, M.P. Potential alkylating agents from the oxidation of carcinogenic cyclic N-nitrosamines. Br. J. Cancer 1973, 28, 84. [Google Scholar] [CrossRef] [Green Version]
- Lijinsky, W.; Taylor, H.W. Carcinogenicity of methylated nitrosopiperidines. Int. J. Cancer 1975, 16, 318–322. [Google Scholar] [CrossRef]
- Lijinsky, W.; Taylor, H.W. Tumorigenesis by oxygenated nitrosopiperidines in rats. J. Natl. Cancer Inst. 1975, 55, 705–708. [Google Scholar] [CrossRef]
- Liu, Z.; Young-Sciame, R.; Hecht, S.S. Liquid chromatography-electrospray ionization mass spectrometric detection of an ethenodeoxyguanosine adduct and its hemiaminal precursors in DNA reacted with α-acetoxy-N-nitrosopiperidine and cis-4-oxo-2-pentenal. Chem. Res. Toxicol. 1996, 9, 774–780. [Google Scholar] [CrossRef]
- Totsuka, Y.; Lin, Y.; He, Y.; Ishino, K.; Sato, H.; Kato, M.; Nagai, M.; Elzawahry, A.; Totoki, Y.; Nakamura, H.; et al. DNA adductome analysis identifies N-nitrosopiperidine involved in the etiology of esophageal cancer in Cixian, China. Chem. Res. Toxicol. 2019, 32, 1515–1527. [Google Scholar] [CrossRef] [Green Version]
- Sakaguchi, S.; Yokokawa, Y.; Hou, J.; Zhang, X.L.; Li, X.P.; Li, S.S.; Li, X.X.; Zhu, D.C.; Kamijima, M.; Yamanoshita, O.; et al. Environmental exposure and p53 mutations in esophageal cancer patients in areas of low and high incidence of esophageal cancer in China. Tohoku J. Exp. Med. 2005, 207, 313–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spiegelhalder, B.; Preussmann, R. Contamination of toiletries and cosmetic products with volatile and nonvolatile N-nitroso carcinogens. J. Cancer Res. Clin. Oncol. 1984, 108, 160–163. [Google Scholar] [CrossRef] [PubMed]
- Sanches Filho, P.J.; Rios, A.; Valcárcel, M.; Zanin, K.D.; Caramão, E.B. Determination of nitrosamines in preserved sausages by solid-phase extraction-micellar electrokinetic chromatography. J. Chromatogr. A 2003, 985, 503–512. [Google Scholar] [CrossRef]
- Charrois, J.W.; Arend, M.W.; Froese, K.L.; Hrudey, S.E. Detecting N-nitrosamines in drinking water at nanogram per liter levels using ammonia positive chemical ionization. Environ. Sci. Technol. 2004, 38, 4835–4841. [Google Scholar] [CrossRef]
- Reh, B.D.; Fajen, J.M. Worker exposures to nitrosamines in a rubber vehicle sealing plant. Am. Ind. Hyg. Assoc. J. 1996, 57, 918–923. [Google Scholar] [CrossRef]
- Fajen, J.M.; Carson, G.A.; Rounbehler, D.P.; Fan, T.Y.; Vita, R.; Goff, U.E.; Wolf, M.H.; Edwards, G.S.; Fine, D.H.; Reinhold, V.; et al. N-nitrosamines in the rubber and tire industry. Science 1979, 205, 1262–1264. [Google Scholar] [CrossRef]
- Oury, B.; Limasset, J.C.; Protois, J.C. Assessment of exposure to carcinogenic N-nitrosamines in the rubber industry. Int. Arch. Occup. Environ. Health 1997, 70, 261–271. [Google Scholar] [CrossRef]
- Monarca, S.; Feretti, D.; Zanardini, A.; Moretti, M.; Villarini, M.; Spiegelhalder, B.; Zerbini, I.; Gelatti, U.; Lebbolo, E. Monitoring airborne genotoxicants in the rubber industry using genotoxicity tests and chemical analyses. Mutat. Res. 2001, 490, 159–169. [Google Scholar] [CrossRef]
- Mirvish, S.S. Formation of N-nitroso compounds: Chemistry, kinetics, and in vivo occurrence. Toxicol. Appl. Pharmacol. 1975, 31, 325–351. [Google Scholar] [CrossRef]
- Hecht, S.S.; Morrison, J.B. A sensitive method for detecting in vivo formation of N-nitrosomorpholine and its application to rats given low doses of morpholine and sodium nitrite. Cancer Res. 1984, 44, 2873–2877. [Google Scholar]
- Van Stee, E.W.; Sloane, R.A.; Simmons, J.E.; Moorman, M.P.; Brunnemann, K.D. Endogenous formation of N-nitrosomorpholine in mice from 15NO2 by inhalation and morpholine by gavage. Carcinogenesis 1995, 16, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Singer, G.M.; Lijinsky, W. Naturally occurring nitrosatable compounds. I. Secondary amines in foodstuffs. J. Agric. Food Chem. 1976, 24, 550–553. [Google Scholar] [CrossRef] [PubMed]
- Börzsönyi, M.; Surján, A.; Pintér, A.; Török, G.; Csik, M.; Tamás, J.; Fetter, J.; Ferencz, A. Endogenous Formation of N-Nitrosomorpholine from a New Fungicide. In IARC Scientific Publication No. 31: N-Nitroso Compounds: Analysis, Formation and Occurrence; Walker, E., Griciute, L., Castegnaro, M., Borzsonyi, M., Eds.; IARC: Lyon, France, 1980; pp. 677–683. [Google Scholar]
- Brunnemann, K.D.; Scott, J.C.; Hoffmann, D. N-Nitrosomorpholine and other volatile N-nitrosamines in snuff tobacco. Carcinogenesis 1982, 3, 693–696. [Google Scholar] [CrossRef] [PubMed]
- McGlothlin, J.D.; Wilcox, T.C.; Fajen, J.M.; Edwards, G.S. A Health Hazard Evaluation of Nitrosamines in a Tire Manufacturing Plant. In Chemical Hazards in the Workplace; American Chemical Society: Washington, DC, USA, 1981; Volume 149, pp. 283–299. [Google Scholar]
- Spiegelhalder, B.; Preussmann, R. Occupational nitrosamine exposure. 1. Rubber and tyre industry. Carcinogenesis 1983, 4, 1147–1152. [Google Scholar] [CrossRef]
- Kakizoe, T.; Wang, T.T.; Eng, V.W.; Furrer, R.; Dion, P.; Bruce, W.R. Volatile N-nitrosamines in the urine of normal donors and of bladder cancer patients. Cancer Res. 1979, 39, 829–832. [Google Scholar]
- van Maanen, J.M.; Pachen, D.M.; Dallinga, J.W.; Kleinjans, J.C. Formation of nitrosamines during consumption of nitrate- and amine-rich foods, and the influence of the use of mouthwashes. Cancer Detect. Prev. 1998, 22, 204–212. [Google Scholar] [CrossRef]
- Dallinga, J.W.; Pachen, D.M.; Lousberg, A.H.; van Geel, J.A.; Houben, G.M.; Stockbrügger, R.W.; van Maanen, J.M.; Kleinjans, J.C. Volatile N-nitrosamines in gastric juice of patients with various conditions of the gastrointestinal tract determined by gas chromatography-mass spectrometry and related to intragastric pH and nitrate and nitrite levels. Cancer Lett. 1998, 124, 119–125. [Google Scholar] [CrossRef]
- Lijinsky, W.; Kovatch, R.M.; Knutsen, G.L. Carcinogenesis by nitrosomorpholines, nitrosooxazolidines and nitrosoazetidine given by gavage to Syrian golden hamsters. Carcinogenesis 1984, 5, 875–878. [Google Scholar] [CrossRef]
- Rao, M.S.; Scarpelli, D.G.; Lijinsky, W. Carcinogenesis in Syrian hamsters by N-nitroso-2,6-dimethylmorpholine, its cis and trans isomers, and the effect of deuterium labeling. Carcinogenesis 1981, 2, 731–735. [Google Scholar] [CrossRef]
- Palladino, G.F.; Chung, F.L.; Hecht, S.S. 3-(2-Deoxy-ß-D-erythropentofuranosyl)-6,7-dihydro-6,7-dihydroxyimidazo[1,2-a]purin-9(3H)-one, a major deoxyguanosine adduct formed from a novel diazo hydroxide product of α-hydroxylation of the carcinogen N-nitrosomorpholine. J. Am. Chem. Soc. 1986, 108, 6066–6068. [Google Scholar] [CrossRef]
- Stewart, B.W.; Swann, P.F.; Holsman, J.W.; Magee, P.N. Cellular injury and carcinogenesis. Evidence for the alkylation of rat liver nucleic acids in vivo by N-nitrosomorpholine. Z. Krebsforsch. Klin. Onkol. Cancer Res. Clin. Oncol. 1974, 82, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hecht, S.S.; Young, R. Metabolic α-hydroxylation of N-nitrosomorpholine and 3,3,5,5-tetradeutero-N-nitrosomorpholine in the F344 rat. Cancer Res. 1981, 41 Pt 1, 5039–5043. [Google Scholar] [PubMed]
- Hecht, S.S.; Morrison, J.B.; Young, R. N-Nitroso(2-hydroxyethyl)glycine, a urinary metabolite of N,N-dinitrosopiperazine with potential utility as a monitor for its formation in vivo from piperazine. Carcinogenesis 1984, 5, 979–981. [Google Scholar] [CrossRef] [PubMed]
- Manson, D.; Cox, P.J.; Jarman, M. Metabolism of N-nitrosomorpholine by the rat in vivo and by rat liver microsomes and its oxidation by the Fenton system. Cell Biol. Int. 1978, 20, 341–354. [Google Scholar] [CrossRef]
- Arimoto-Kobayashi, S.; Hayatsu, H. Formation of direct-acting mutagens from mixtures of N-nitrosomorpholine and carboxylates by UVA irradiation. Environ. Mol. Mutagen. 1998, 31, 163–168. [Google Scholar] [CrossRef]
- Chung, F.L.; Hecht, S.S. Formation of the cyclic 1,N2-glyoxal-deoxyguanosine adduct upon reaction of N-nitroso-2-hydroxymorpholine with deoxyguanosine. Carcinogenesis 1985, 6, 1671–1673. [Google Scholar] [CrossRef] [PubMed]
- Zink, C.N.; Kim, H.J.; Fishbein, J.C. Synthesis and aqueous chemistry of α-acetoxy-N-nitrosomorpholine: Reactive intermediates and products. J. Org. Chem. 2006, 71, 202–209. [Google Scholar] [CrossRef]
- Zink, C.N.; Soissons, N.; Fishbein, J.C. Products of the direct reaction of the diazonium ion of a metabolite of the carcinogen N-nitrosomorpholine with purines of nucleosides and DNA. Chem. Res. Toxicol. 2010, 23, 1223–1233. [Google Scholar] [CrossRef]
- Kim, H.J.; Fishbein, J.C. Reexamination of the aqueous chemistry of N-nitroso-3-hydroxymorpholine, a metabolite of the carcinogen N-nitrosomorpholine. Chem. Res. Toxicol. 2003, 16, 715–720. [Google Scholar] [CrossRef]
- Koissi, N.; Shah, N.H.; Ginevan, B.; Eck, W.S.; Roebuck, B.D.; Fishbein, J.C. Lactone metabolite common to the carcinogens dioxane, diethylene glycol, and N-nitrosomorpholine: Aqueous chemistry and failure to mediate liver carcinogenesis in the F344 rat. Chem. Res. Toxicol. 2012, 25, 1022–1028. [Google Scholar] [CrossRef] [Green Version]
- Eisenbrand, G.; Denkel, E.; Pool, B. Alcoholdehydrogenase as an activating enzyme for N-nitrosodiethanolamine (NDELA): In vitro activation of NDELA to a potent mutagen in Salmonella typhimurium. J. Cancer Res. Clin. Oncol. 1984, 108, 76–80. [Google Scholar] [CrossRef] [PubMed]
- Denkel, E.; Pool, B.L.; Schlehofer, J.R.; Eisenbrand, G. Biological activity of N-nitrosodiethanolamine and of potential metabolites which may arise after activation by alcohol dehydrogenase in Salmonella typhimurium, in mammalian cells, and in vivo. J. Cancer Res. Clin. Oncol. 1986, 111, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Koissi, N.; Fishbein, J.C. Trapping of a cross-link formed by a major purine adduct of a metabolite of the carcinogen N-nitrosomorpholine by inorganic and biological reductants. Chem. Res. Toxicol. 2013, 26, 732–740. [Google Scholar] [CrossRef] [Green Version]
- Hecht, S.S. Approaches to cancer prevention based on an understanding of N-nitrosamine carcinogenesis. Proc. Soc. Exp. Biol. Med. 1997, 216, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Ricker, K.; Tsai, F.C.; Hsieh, C.J.; Osborne, G.; Sun, M.; Marder, M.E.; Elmore, S.; Schmitz, R.; Sandy, M.S. Estimated cancer risks associated with nitrosamine contamination in commonly used medications. Int. J. Environ. Res. Public Health 2021, 18, 9465. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [Green Version]
- Balbo, S.; Turesky, R.J.; Villalta, P.W. DNA adductomics. Chem. Res. Toxicol. 2014, 27, 356–366. [Google Scholar] [CrossRef]


























| Animal Species | Administration Pathway | Exposure Amount | Exposure Time | Target Tissue | DNA Adducts (μmol/mol dGuo unless otherwise Noted) Formed by NPYR | Ref. | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N7,8-Butanoguanine | N7-(4-Oxobutyl)-dGuo | N1,N2-Propano-Gua a | N6-(4-HOB)-dAdo b | N2-(4-HOB)-dGuo b | O2-(4-HOB)-Thd b | O4-(4-HOB)-Thd b | ||||||
| Male F344 rats | Single dose by gavage | 3.1 mg/kg (8 mCi) [3,4-3H]NPYR | 16 h | Liver | 0.7 pmol/mg DNA | [244] | ||||||
| 6.7 mg/kg (15 mCi) [3,4-3H]NPYR | ND | |||||||||||
| 175 mg/kg NPYR | 320 pmol/mg DNA | |||||||||||
| 350 mg/kg NPYR | 390 pmol/mg DNA | |||||||||||
| 450 mg/kg NPYR | 24 h | Liver RNA | 2860 ± 332 | ND | [249] | |||||||
| Liver DNA | 1430 ± 56 | 603 | ||||||||||
| 46 mg/kg NPYR | 16 h | Liver | 952 ± 180 | 0.231 ± 0.078 | 0.013 ± 0.006 | 0.369 ± 0.134 | 1.065 ± 0.349 | [247] | ||||
| 92 mg/kg NPYR | 1742 ± 245 | 0.499 ± 0.091 | 0.008 ± 0.004 | 0.478 ± 0.114 | 2.049 ± 0.361 | |||||||
| 184 mg/kg NPYR | 3032 ± 855 | 0.905 ± 0.240 | 0.016 ± 0.008 | 0.951 ± 0.193 | 3.767 ± 0.840 | |||||||
| Single dose by i.p. injection | 450 mg/kg NPYR | 16 h | Liver | 1792 ± 87 | 643 ± 9 | ND | [250] | |||||
| In the drinking water | 6 mM NPYR | 14 days | Liver | 0.06 | [251] | |||||||
| 600 ppm NPYR | 1 week | 0.02 ± 0.01 | 3.41 ± 0.60 | 2.56 ± 0.84 | 2.28 ± 0.47 | [252] | ||||||
| 200 ppm NPYR | 4 weeks | 0.03 ± 0.01 | 4.84 ± 0.48 | 3.87 ± 0.97 | 3.83 ± 0.57 | |||||||
| 200 ppm NPYR | 13 weeks | 0.04 ± 0.02 | 5.39 ± 0.39 | 3.52 ± 0.43 | 5.05 ± 1.68 | |||||||
| Male Sprague-Dawley rats | Single dose by intubation | 14–900 mg/kg NPYR | 12 h | Liver | Dose-dependently formed; 0.1–3 equivalents of N7-Me-Gua | [243] | ||||||
| 0.63 mg [14C]NPYR/ 1 rat (207 g) | 8- or 42.6-fold of N7-Me-Gua radioactivity | |||||||||||
| Intragastric administration | 900 mg/kg NPYR | 12 h | Liver | 3983 | [248] | |||||||
| Kidney | 159 | |||||||||||
| Lung | 159 | |||||||||||
| Male Swiss Webster mice | Intragastric administration | 900 mg/kg NPYR | 24 h | Liver | 9987 | [248] | ||||||
| Kidney | 699 | |||||||||||
| Lung | 499 | |||||||||||
| Male Syrian golden hamsters | Intragastric administration | 900 mg/kg NPYR | 24 h | Liver | 9000 | [248] | ||||||
| Kidney | 900 | |||||||||||
| Lung | 1170 | |||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Hecht, S.S. Metabolic Activation and DNA Interactions of Carcinogenic N-Nitrosamines to Which Humans Are Commonly Exposed. Int. J. Mol. Sci. 2022, 23, 4559. https://doi.org/10.3390/ijms23094559
Li Y, Hecht SS. Metabolic Activation and DNA Interactions of Carcinogenic N-Nitrosamines to Which Humans Are Commonly Exposed. International Journal of Molecular Sciences. 2022; 23(9):4559. https://doi.org/10.3390/ijms23094559
Chicago/Turabian StyleLi, Yupeng, and Stephen S. Hecht. 2022. "Metabolic Activation and DNA Interactions of Carcinogenic N-Nitrosamines to Which Humans Are Commonly Exposed" International Journal of Molecular Sciences 23, no. 9: 4559. https://doi.org/10.3390/ijms23094559
APA StyleLi, Y., & Hecht, S. S. (2022). Metabolic Activation and DNA Interactions of Carcinogenic N-Nitrosamines to Which Humans Are Commonly Exposed. International Journal of Molecular Sciences, 23(9), 4559. https://doi.org/10.3390/ijms23094559