Alzheimer’s Disease: Causal Effect between Obesity and APOE Gene Polymorphisms
Abstract
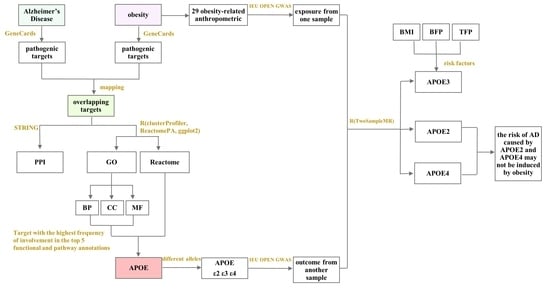
:1. Introduction
2. Results
2.1. Bioinformatics Analysis
2.1.1. Obesity- and AD-Related Targets, and PPI Network of Overlapping Targets
2.1.2. Functional and Pathway Annotations of Overlapping Targets between Obesity and AD
Biological Processes to GO Enrichment Analysis Results of Overlapping Targets between Obesity and AD
Cellular Components to GO Enrichment Analysis Results of Overlapping Targets between Obesity and AD
Molecular Functions to GO Enrichment Analysis Results of Overlapping Targets between Obesity and AD
Reactome Pathways of Overlapping Targets between Obesity and AD
Frequency of Involvement of Overlapping Targets between Obesity and AD in Top Five Functional and Pathway Annotations
2.2. Mendelian Randomization Analysis
2.2.1. Causal Association between Different Types of Body Anthropometric Indexes of Obesity and APOE2
2.2.2. Causal Association between Different Types of Body Anthropometric Indexes of Obesity and APOE3
2.2.3. Causal Association between Different Types of Body Anthropometric Indexes of Obesity and APOE4
3. Discussion
4. Methods and Materials
4.1. Databases and Software
4.2. Bioinformatics and Mendelian Randomization Analysis of the Mechanism of Obesity-Induced AD
4.3. Bioinformatics Analysis
4.3.1. Screening of Obesity-Related Targets
4.3.2. Screening of AD-Related Targets
4.3.3. Protein–Protein Interaction (PPI) Network of Overlapping Targets between Obesity and AD
4.3.4. GO Enrichment and Reactome Pathway Analyses of Overlapping Targets between Obesity and AD
4.4. Mendelian Randomization (MR) Analysis
4.4.1. Data Source
4.4.2. Selection of Instrumental Variables
4.4.3. Statistical Inference for Causal Effects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 2SMR | two-sample Mendelian randomization |
| AD | Alzheimer’s disease |
| ALB | Albumin |
| APOA1 | Apolipoprotein A-I |
| APOB | Apolipoprotein B-100 |
| APOE | Apolipoprotein E |
| AS | astrocytes |
| Aβ | amyloid β-protein |
| BBB | blood–brain barrier |
| BFP | body fat percentage |
| BMI | body mass index |
| BP | biological process |
| CC | cellular component |
| CETP | cholesteryl ester transfer protein |
| CI | confidence interval |
| CNS | central nervous system |
| GO | gene ontology |
| GWAS | genome-wide association study |
| HDL | high density lipoprotein |
| IL18 | Interleukin 18 |
| IL1B | Interleukin 1 beta |
| IL6 | Interleukin 6 |
| INS | insulin |
| IVs | instrumental variables |
| IVW | inverse variance Weighted |
| LD | linkage disequilibrium |
| LPL | lipoprotein lipase |
| MF | molecular function |
| MR | Mendelian randomization |
| OR | odds ratio |
| PON1 | serum paraoxonase/arylesterase 1 |
| PPI | protein–protein interaction |
| PTEN | Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN |
| RCT | randomized controlled trial |
| SD | Standard Deviation |
| SNPs | single-nucleotide polymorphisms |
| TFP | trunk fat percentage |
| TNF | tumor necrosis factors |
| VLDL | very low-density lipoprotein |
| WHO | World Health Organization |
References
- Alzheimer’s Association. 2022 Alzheimer’s disease facts and figures. Alzheimer’s Dement. J. Alzheimer’s Assoc. 2002, 18, 700–789. [Google Scholar]
- Naderali, E.K.; Ratcliffe, S.H.; Dale, M.C. Obesity and Alzheimer’s disease: A link between body weight and cognitive function in old age. Am. J. Alzheimer’s Dis. Other Dement. 2009, 24, 445–449. [Google Scholar] [CrossRef]
- Morys, F.; Potvin, O.; Zeighami, Y.; Vogel, J.; Lamontagne-Caron, R.; Duchesne, S.; Dagher, A. Obesity-Associated Neurodegeneration Pattern Mimics Alzheimer’s Disease in an Observational Cohort Study. J. Alzheimer’s Dis. 2022, 91, 1059–1071. [Google Scholar] [CrossRef]
- Profenno, L.A.; Porsteinsson, A.P.; Faraone, S.V. Meta-analysis of Alzheimer’s disease risk with obesity, diabetes, and related disorders. Biol. Psychiatry 2010, 67, 505–512. [Google Scholar] [CrossRef]
- Whitmer, R.A.; Gunderson, E.P.; Barrett-Connor, E.; Quesenberry, C.P.; Yaffe, K. Obesity in middle age and future risk of dementia: A 27-year longitudinal population based study. Br. Med. J. 2005, 330, 1360. [Google Scholar] [CrossRef]
- Beydoun, M.A.; Beydoun, H.A.; Wang, Y. Obesity and central obesity as risk factors for incident dementia and its subtypes: A systematic review and meta-analysis. Obes. Rev. Off. J. Int. Assoc. Study Obes. 2008, 9, 204–218. [Google Scholar] [CrossRef]
- Berlinger, W.G.; Potter, J.F. Low Body Mass Index in demented outpatients. J. Am. Geriatr. Soc. 1991, 39, 973–978. [Google Scholar] [CrossRef]
- Burns, A.; Marsh, A.; Bender, D.A. Dietary intake and clinical, anthropometric and biochemical indices of malnutrition in elderly demented patients and non-demented subjects. Psychol. Med. 1989, 19, 383–391. [Google Scholar] [CrossRef]
- Nourhashemi, F.; Deschamps, V.; Larrieu, S.; Letenneur, L.; Dartigues, J.-F.; Barberger-Gateau, P. Body mass index and incidence of dementia: The PAQUID study. Neurology 2003, 60, 117–119. [Google Scholar] [CrossRef]
- Qu, S.; Lu, H.; Song, Y.F. Multidisciplinary Clinical Consensus on Diagnosis and Treatment of Obesity (2021 Edition). Chin. J. Obes. Metab. 2021, 7, 211–226. Available online: https://kns.cnki.net/kcms2/article/abstract?v=3uoqIhG8C44YLTlOAiTRKibYlV5Vjs7iJTKGjg9uTdeTsOI_ra5_XQ9BXpo0MMlqxGYwMG8ixqM5JbD8Rm7lkCs5dPl5ON7h&uniplatform=NZKPT (accessed on 18 February 2023).
- Deng, Y.-T.; Li, Y.-Z.; Huang, S.-Y.; Ou, Y.-N.; Zhang, W.; Chen, S.-D.; Zhang, Y.-R.; Yang, L.; Dong, Q.; Feng, J.-F.; et al. Association of life course adiposity with risk of incident dementia: A prospective cohort study of 322,336 participants. Mol. Psychiatry 2022, 27, 3385–3395. [Google Scholar] [CrossRef]
- Jagust, W.; Harvey, D.; Mungas, D.; Haan, M. Central obesity and the aging brain. Arch. Neurol. 2005, 62, 1545–1548. [Google Scholar] [CrossRef]
- Tang, X.; Zhao, W.; Lu, M.; Zhang, X.; Zhang, P.; Xin, Z.; Sun, R.; Tian, W.; Cardoso, M.A.; Yang, J.; et al. Relationship between Central Obesity and the incidence of Cognitive Impairment and Dementia from Cohort Studies Involving 5,060,687 Participants. Neurosci. Biobehav. Rev. 2021, 130, 301–313. [Google Scholar] [CrossRef]
- Gustafson, D.R.; Backman, K.; Waern, M.; Ostling, S.; Guo, X.; Zandi, P.; Mielke, M.M.; Bengtsson, C.; Skoog, I. Adiposity indicators and dementia over 32 years in Sweden. Neurology 2009, 73, 1559–1566. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, A.L.; Kuller, L.H.; Lopez, O.L.; Diehr, P.; O’Meara, E.S.; Longstreth, W.T.; Luchsinger, J.A. Midlife and late-life obesity and the risk of dementia: Cardiovascular health study. Arch. Neurol. 2009, 66, 336–342. [Google Scholar] [CrossRef]
- Jayedi, A.; Soltani, S.; Zargar, M.S.; Khan, T.A.; Shab-Bidar, S. Central fatness and risk of all-cause mortality: Systematic review and dose-response meta-analysis of 72 prospective cohort studies. Br. Med. J. 2020, 370, m3324. [Google Scholar] [CrossRef] [PubMed]
- Obesity: Preventing and managing the global epidemic. In Report of a WHO Consultation; World Health Organization technical report series; World Health Organization: Geneve, Switzerland, 2000; Volume 894, pp. 1–253.
- Huang, S.-Y.; Yang, Y.-X.; Chen, S.-D.; Li, H.-Q.; Zhang, X.-Q.; Kuo, K.; Tan, L.; Feng, L.; Dong, Q.; Zhang, C.; et al. Investigating causal relationships between exposome and human longevity: A Mendelian randomization analysis. BMC Med. 2021, 19, 150. [Google Scholar] [CrossRef]
- Shinohara, M.; Kanekiyo, T.; Yang, L.; Linthicum, D.; Shinohara, M.; Fu, Y.; Price, L.; Frisch-Daiello, J.L.; Han, X.; Fryer, J.D.; et al. APOE2 eases cognitive decline during Aging: Clinical and preclinical evaluations. Ann. Neurol. 2016, 79, 758–774. [Google Scholar] [CrossRef]
- Zhao, L.; Gottesdiener, A.J.; Parmar, M.; Li, M.; Kaminsky, S.M.; Chiuchiolo, M.J.; Sondhi, D.; Sullivan, P.M.; Holtzman, D.M.; Crystal, R.G.; et al. Intracerebral adeno-associated virus gene delivery of apolipoprotein E2 markedly reduces brain amyloid pathology in Alzheimer’s disease mouse models. Neurobiol. Aging 2016, 44, 159–172. [Google Scholar] [CrossRef] [PubMed]
- Sepulveda-Falla, D.; Sanchez, J.S.; Almeida, M.C.; Boassa, D.; Acosta-Uribe, J.; Vila-Castelar, C.; Ramirez-Gomez, L.; Baena, A.; Aguillon, D.; Villalba-Moreno, N.D.; et al. Distinct tau neuropathology and cellular profiles of an APOE3 Christchurch homozygote protected against autosomal dominant Alzheimer’s dementia. Acta Neuropathol. 2022, 144, 589–601. [Google Scholar] [CrossRef]
- Mahley, R.W.; Huang, Y. Apolipoprotein e sets the stage: Response to injury triggers neuropathology. Neuron 2012, 76, 871–885. [Google Scholar] [CrossRef]
- Liu, C.-C.; Kanekiyo, T.; Xu, H.; Bu, G. Apolipoprotein E and Alzheimer disease: Risk, mechanisms and therapy. Nat. Rev. Neurol. 2013, 9, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Weisgraber, K.H.; Mucke, L.; Mahley, R.W. Apolipoprotein E: Diversity of cellular origins, structural and biophysical properties, and effects in Alzheimer’s disease. J. Mol. Neurosci. 2004, 23, 189–204. [Google Scholar] [CrossRef]
- Linton, M.F.; Gish, R.; Hubl, S.T.; Bütler, E.; Esquivel, C.; I Bry, W.; Boyles, J.K.; Wardell, M.R.; Young, S.G. Phenotypes of apolipoprotein B and apolipoprotein E after liver transplantation. J. Clin. Investig. 1991, 88, 270–281. [Google Scholar] [CrossRef] [PubMed]
- Davies, N.M.; Holmes, M.V.; Davey Smith, G. Reading Mendelian Randomisation Studies: A Guide, Glossary, and Checklist for Clinicians. BMJ 2018, 362, k601. Available online: https://pubmed.ncbi.nlm.nih.gov/30002074/ (accessed on 6 February 2023).
- Sparks, D.; Liu, H.; Gross, D.R.; Scheff, S.W. Increased density of cortical apolipoprotein E immunoreactive neurons in rabbit brain after dietary administration of cholesterol. Neurosci. Lett. 1995, 187, 142–144. [Google Scholar] [CrossRef]
- Weisgraber, K.H. Apolipoprotein E distribution among human plasma lipoproteins: Role of the cysteine-arginine interchange at residue 112. J. Lipid Res. 1990, 31, 1503–1511. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, J.; Kouiavskaia, D.; Migliorini, M.; Robinson, S.; Saenko, E.L.; Gorlatova, N.; Li, D.; Lawrence, D.; Hyman, B.T.; Weisgraber, K.H.; et al. The apoE isoform binding properties of the VLDL receptor reveal marked differences from LRP and the LDL receptor. J. Lipid Res. 2005, 46, 1721–1731. [Google Scholar] [CrossRef]
- Hatters, D.M.; Zhong, N.; Rutenber, E.; Weisgraber, K.H. Amino-terminal domain stability mediates apolipoprotein E aggregation into neurotoxic fibrils. J. Mol. Biol. 2006, 361, 932–944. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Painter, M.M.; Bu, G.; Kanekiyo, T. Apolipoprotein E as a Therapeutic Target in Alzheimer’s Disease: A Review of Basic Research and Clinical Evidence. CNS Drugs 2016, 30, 773–789. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Zhao, Z.; Sagare, A.P.; Wu, Y.; Wang, M.; Owens, N.C.; Verghese, P.B.; Herz, J.; Holtzman, D.M.; Zlokovic, B.V. Blood-Brain Barrier-Associated Pericytes Internalize and Clear Aggregated amyloid-β42 by LRP1-Dependent Apolipoprotein E Isoform-Specific Mechanism. Mol. Neurodegener. 2018, 13, 57. Available online: https://pubmed.ncbi.nlm.nih.gov/30340601/ (accessed on 8 February 2023).
- Liu, L.; MacKenzie, K.R.; Putluri, N.; Maletić-Savatić, M.; Bellen, H.J. The Glia-Neuron Lactate Shuttle and Elevated ROS Promote Lipid Synthesis in Neurons and Lipid Droplet Accumulation in Glia via APOE/D. Cell Metab. 2017, 26, 719–737.e6. [Google Scholar] [CrossRef] [PubMed]
- Osenkowski, P.; Ye, W.; Wang, R.; Wolfe, M.S.; Selkoe, D.J. Direct and potent regulation of gamma-secretase by its lipid microenvironment. J. Biol. Chem. 2008, 283, 22529–22540. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2006, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Zhang, H.; Liu, Y.; Liu, Y.-X.; Huang, L. EVenn: Easy to create repeatable and editable Venn diagrams and Venn networks online. J. Genet. Genom. 2021, 48, 863–866. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Hemani, G.; Zheng, J.; Elsworth, B.; Wade, K.H.; Haberland, V.; Baird, D.; Laurin, C.; Burgess, S.; Bowden, J.; Langdon, R.; et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018, 7, e34408. [Google Scholar] [CrossRef]
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 4 February 2022).
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef] [PubMed]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020, 48, D498–D503. [Google Scholar] [CrossRef]
- Smith, G.D.; Hemani, G. Mendelian randomization: Genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 2014, 23, R89–R98. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.D.; Ebrahim, S. Mendelian randomization: Can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003, 32, 1–22. [Google Scholar] [CrossRef]
- Emdin, C.A.; Khera, A.V.; Kathiresan, S. Mendelian Randomization. JAMA 2017, 318, 1925–1926. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Zhao, N.; Caulfield, T.R.; Liu, C.-C.; Bu, G. Apolipoprotein E and Alzheimer disease: Pathobiology and targeting strategies. Nat. Clin. Pract. Neurol. 2019, 15, 501–518. [Google Scholar] [CrossRef]
- Bowden, J.; Del Greco, M.F.; Minelli, C.; Davey Smith, G.; Sheehan, N.; Thompson, J. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat. Med. 2017, 36, 1783–1802. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Thompson, S.G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 2017, 32, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Verbanck, M.; Chen, C.-Y.; Neale, B.; Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018, 50, 693–698. [Google Scholar] [CrossRef] [PubMed]







| IVW-β (95%CI) | APOE2 (4% AD) | APOE3 (58% AD) | APOE4 (38% AD) |
|---|---|---|---|
| body mass index | −0.205(−0.371,−0.040) | −0.194(−0.351,−0.038) | / |
| Weight | −0.201(−0.356,−0.046) | −0.211(−0.365,−0.057) | / |
| Body fat percentage | / | −0.302(−0.537,−0.066) | / |
| Whole body fat mass | −0.205(−0.371,−0.038) | −0.224(−0.387,−0.061) | / |
| Whole body fat-free mass | / | / | −1.255(−2.312,−0.199) |
| Whole body water mass | / | / | −1.350(−2.347,−0.353) |
| Arm fat mass (right) | / | −0.178(−0.338,−0.018) | / |
| Arm fat mass (left) | / | −0.190(−0.350,−0.030) | / |
| Arm fat-free mass (right) | / | −0.261(−0.462,−0.059) | −1.422(−2.352,−0.492) |
| Arm fat-free mass (left) | / | −0.229(−0.426,−0.032) | −1.455(−2.570,−0.340) |
| Leg fat mass (right) | / | −0.242(−0.441,−0.042) | / |
| Leg fat mass (left) | / | −0.245(−0.447,−0.043) | / |
| Leg fat-free mass (left) | −0.199(−0.392,−0.005) | −0.298(−0.489,−0.106) | / |
| Trunk fat percentage | / | −0.275(−0.473,−0.078) | / |
| Trunk fat mass | −0.216(−0.383,−0.049) | −0.261(−0.425,−0.098) | / |
| Exposure Trait | GWAS ID | Sample Size | Number of SNPs | Consortium | PMID | Sex | Year |
|---|---|---|---|---|---|---|---|
| Body mass index | ieu-b-40 | 681,275 | 2,336,260 | GIANT | 30124842 | Males and Females | 2018 |
| Weight | ukb-b-11842 | 461,632 | 9,851,867 | MRC-IEU | — | Males and Females | 2018 |
| Body fat | ieu-a-999 | 100,716 | 3,228,665 | — | 26833246 | Males and Females | 2016 |
| Body fat percentage | ukb-b-8909 | 454,633 | 9,851,867 | MRC-IEU | — | Males and Females | 2018 |
| Whole body fat mass | ukb-b-19393 | 454,137 | 9,851,867 | MRC-IEU | — | Males and Females | 2018 |
| Whole body fat-free mass | ukb-b-13354 | 454,850 | 9,851,867 | MRC-IEU | — | Males and Females | 2018 |
| Whole body water mass | ukb-b-14540 | 454,888 | 9,851,867 | MRC-IEU | — | Males and Females | 2018 |
| Appendicular lean mass | ebi-a-GCST90000025 | 450,243 | 18,071,518 | — | 33097823 | Males and Females | 2020 |
| Arm fat percentage (right) | ukb-b-12854 | 454,789 | 9,851,867 | MRC-IEU | — | Males and Females | 2018 |
| Arm fat percentage (left) | ukb-b-20188 | 454,724 | 9,851,867 | MRC-IEU | — | Males and Females | 2018 |
| Arm fat mass (right) | ukb-b-6704 | 454,757 | 9,851,867 | MRC-IEU | — | Males and Females | 2018 |
| Arm fat mass (left) | ukb-b-8338 | 454,684 | 9,851,867 | MRC-IEU | — | Males and Females | 2018 |
| Arm fat-free mass (right) | ukb-b-19520 | 454,753 | 9,851,867 | MRC-IEU | — | Males and Females | 2018 |
| Arm fat-free mass (left) | ukb-b-19925 | 454,672 | 9,851,867 | MRC-IEU | — | Males and Females | 2018 |
| Leg fat percentage (right) | ukb-b-20531 | 454,854 | 9,851,867 | MRC-IEU | — | Males and Females | 2018 |
| Leg fat percentage (left) | ukb-b-18377 | 454,826 | 9,851,867 | MRC-IEU | — | Males and Females | 2018 |
| Leg fat mass (right) | ukb-b-18096 | 454,846 | 9,851,867 | MRC-IEU | — | Males and Females | 2018 |
| Leg fat mass (left) | ukb-b-7212 | 454,823 | 9,851,867 | MRC-IEU | — | Males and Females | 2018 |
| Leg fat-free mass (right) | ukb-b-12828 | 454,835 | 9,851,867 | MRC-IEU | — | Males and Females | 2018 |
| Leg fat-free mass (left) | ukb-b-16099 | 454,805 | 9,851,867 | MRC-IEU | — | Males and Females | 2018 |
| Hip circumference | ieu-a-49 | 213,038 | 2,559,739 | GIANT | 25673412 | Males and Females | 2015 |
| Hip circumference adjusted for BMI | ieu-a-55 | 211,114 | 2,540,926 | GIANT | 25673412 | Males and Females | 2015 |
| Waist circumference | ieu-a-61 | 232,101 | 2,565,408 | GIANT | 25673412 | Males and Females | 2015 |
| Waist circumference adjusted for BMI | ieu-a-67 | 231,353 | 2,546,074 | GIANT | 25673412 | Males and Females | 2015 |
| Waist-to-hip ratio | ieu-a-73 | 212,244 | 2,560,782 | GIANT | 25673412 | Males and Females | 2015 |
| Waist-to-hip ratio Adjusted for BMI | ieu-a-79 | 210,082 | 2,542,432 | GIANT | 25673412 | Males and Females | 2015 |
| Trunk fat percentage | ukb-b-16407 | 454,613 | 9,851,867 | MRC-IEU | — | Males and Females | 2018 |
| Trunk fat mass | ukb-b-20044 | 454,588 | 9,851,867 | MRC-IEU | — | Males and Females | 2018 |
| Trunk fat-free mass | ukb-b-17409 | 454,508 | 9,851,867 | MRC-IEU | — | Males and Females | 2018 |
| Outcome Trait | GWAS ID | Sample Size | Number of SNPs | Consortium | PMID | Sex | Year |
|---|---|---|---|---|---|---|---|
| Apolipoprotein E (isoform E2) | prot-a-132 | 3301 | 10,534,735 | — | 29875488 | Males and Females | 2018 |
| Apolipoprotein E (isoform E3) | prot-a-131 | 3301 | 10,534,735 | — | 29875488 | Males and Females | 2018 |
| Apo E4 | prot-c-2938_55_2 | — | 501,428 | — | 28240269 | Males and Females | 2019 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, T.; Zhong, T.; Zhang, M.; Xu, Y.; Zhang, M.; Chen, L. Alzheimer’s Disease: Causal Effect between Obesity and APOE Gene Polymorphisms. Int. J. Mol. Sci. 2023, 24, 13531. https://doi.org/10.3390/ijms241713531
Zhao T, Zhong T, Zhang M, Xu Y, Zhang M, Chen L. Alzheimer’s Disease: Causal Effect between Obesity and APOE Gene Polymorphisms. International Journal of Molecular Sciences. 2023; 24(17):13531. https://doi.org/10.3390/ijms241713531
Chicago/Turabian StyleZhao, Tianyu, Tangsheng Zhong, Meishuang Zhang, Yang Xu, Ming Zhang, and Li Chen. 2023. "Alzheimer’s Disease: Causal Effect between Obesity and APOE Gene Polymorphisms" International Journal of Molecular Sciences 24, no. 17: 13531. https://doi.org/10.3390/ijms241713531
APA StyleZhao, T., Zhong, T., Zhang, M., Xu, Y., Zhang, M., & Chen, L. (2023). Alzheimer’s Disease: Causal Effect between Obesity and APOE Gene Polymorphisms. International Journal of Molecular Sciences, 24(17), 13531. https://doi.org/10.3390/ijms241713531