Machine Learning as a Support for the Diagnosis of Type 2 Diabetes
Abstract
:1. Introduction
2. Results
2.1. Dataset Statistics
2.2. Hyperparameters’ Tuning
- Learning rate: 0.001;
- Loss Function: Binary cross-entropy;
- Optimization algorithm: Stochastic Gradient Descent;
- Trigger function for hidden layer: ReLU;
- Trigger function for the output layer: sigmoids;
2.3. Model Ensemble
2.4. Feature Reduction
2.5. Validation
2.6. Calibration
3. Discussion
4. Materials and Methods
4.1. Features
4.2. Datasets
4.3. Preprocessing
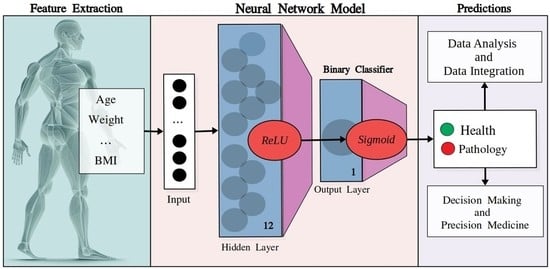
4.4. Neural Networks: Model’s Architecture
4.5. Validation
4.6. Calibration Plots
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- American Diabetes Association Professional Practice Committee. 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes—2022. Diabetes Care 2022, 45 (Suppl. S1), S17–S38. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Saeedi, P.; Karuranga, S.; Pinkepank, M.; Ogurtsova, K.; Duncan, B.B.; Stein, C.; Basit, A.; Chan, J.C.N.; Mbanya, J.C.; et al. IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 2022, 183, 109119. [Google Scholar] [CrossRef] [PubMed]
- International Diabetes Federation. IDF Diabetes Atlas, 10th ed.; International Diabetes Federation: Brussels, Belgium, 2021; Available online: https://www.diabetesatlas.org (accessed on 15 February 2023).
- Liu, P.R.; Lu, L.; Zhang, J.Y.; Huo, T.T.; Liu, S.X.; Ye, Z.W. Application of Artificial Intelligence in Medicine: An Overview. Curr. Med. Sci. 2021, 41, 1105–1115. [Google Scholar] [CrossRef] [PubMed]
- Chicco, D.; Heider, D.; Facchiano, A. Editorial: Artificial Intelligence Bioinformatics: Development and Application of Tools for Omics and Inter-Omics Studies. Front. Genet. 2020, 11, 309. [Google Scholar] [CrossRef]
- Chicco, D.; Facchiano, A.; Tavazzi, E.; Longato, E.; Vettoretti, M.; Bernasconi, A.; Avesani, S.; Cazzaniga, P. (Eds.) Computational Intelligence Methods for Bioinformatics and Biostatistics. In Proceedings of the 17th International Meeting, CIBB 2021, Virtual Event, 15–17 November 2021; Springer: Cham, Switzerland, 2022. Available online: https://link.springer.com/book/10.1007/978-3-031-20837-9 (accessed on 15 February 2023).
- Sheng, B.; Chen, X.; Li, T.; Ma, T.; Yang, Y.; Bi, L.; Zhang, X. An overview of artificial intelligence in diabetic retinopathy and other ocular diseases. Front. Public Health 2022, 10, 971943. [Google Scholar] [CrossRef]
- Ellahham, S. Artificial Intelligence: The Future for Diabetes Care. Am. J. Med. 2020, 133, 895–900. [Google Scholar] [CrossRef]
- Balasubramaniyan, S.; Jeyakumar, V.; Nachimuthu, D.S. Panoramic tongue imaging and deep convolutional machine learning model for diabetes diagnosis in humans. Sci. Rep. 2022, 12, 186. [Google Scholar] [CrossRef]
- Zhang, K.; Liu, X.; Xu, J.; Yuan, J.; Cai, W.; Chen, T.; Wang, K.; Gao, Y.; Nie, S.; Xu, X.; et al. Deep-learning models for the detection and incidence prediction of chronic kidney disease and type 2 diabetes from retinal fundus images. Nat. Biomed. Eng. 2021, 5, 533–545. [Google Scholar] [CrossRef]
- Tang, Y.; Gao, R.; Lee, H.H.; Wells, Q.S.; Spann, A.; Terry, J.G.; Carr, J.J.; Huo, Y.; Bao, S.; Landman, B.A. Prediction of type II diabetes onset with computed tomography and electronic medical records. In Multimodal Learning for Clinical Decision Support and Clinical Image-Based Procedures; Lecture Notes in Computer Science; CLIP ML-CDS 2020 2020; Springer: Cham, Switzerland, 2020; pp. 13–23. [Google Scholar] [CrossRef]
- Dietz, B.; Machann, J.; Agrawal, V.; Heni, M.; Schwab, P.; Dienes, J.; Reichert, S.; Birkenfeld, A.L.; Haring, H.U.; Schick, F.; et al. Detection of diabetes from whole-body MRI using deep learning. JCI Insight 2021, 6, e146999. [Google Scholar] [CrossRef]
- Nomura, A.; Noguchi, M.; Kometani, M.; Furukawa, K.; Yoneda, T. Artificial Intelligence in Current Diabetes Management and Prediction. Curr. Diab. Rep. 2021, 21, 61. [Google Scholar] [CrossRef]
- Ravaut, M.; Harish, V.; Sadeghi, H.; Leung, K.K.; Volkovs, M.; Kornas, K.; Watson, T.; Poutanen, T.; Rosella, L.C. Development and Validation of a Machine Learning Model Using Administrative Health Data to Predict Onset of Type 2 Diabetes. JAMA Netw. Open 2021, 4, e2111315. [Google Scholar] [CrossRef]
- Rein, M.; Ben-Yacov, O.; Godneva, A.; Shilo, S.; Zmora, N.; Kolobkov, D.; Cohen-Dolev, N.; Wolf, B.-C.; Kosower, N.; Lotan-Pompan, M.; et al. Effects of personalized diets by prediction of glycemic responses on glycemic control and metabolic health in newly diagnosed T2DM: A randomized dietary intervention pilot trial. BMC Med. 2022, 20, 56. [Google Scholar] [CrossRef]
- Pavlovskii, V.V.; Derevitskii, I.V.; Kovalchuk, S.V. Hybrid genetic predictive modeling for finding optimal multipurpose multicomponent therapy. J. Comput. Sci. 2022, 63, 101772. [Google Scholar] [CrossRef]
- Murphree, D.H.; Arabmakki, E.; Ngufor, C.; Storlie, C.B.; McCoy, R.G. Stacked classifiers for individualized prediction of glycemic control following initiation of metformin therapy in type 2 diabetes. Comput. Biol. Med. 2018, 103, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Abràmoff, M.D.; Lavin, P.T.; Birch, M.; Shah, N.; Folk, J.C. Pivotal trial of an autonomous AI-based diagnostic system for detection of diabetic retinopathy in primary care offices. NPJ Digit. Med. 2018, 1, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yap, M.H.; Chatwin, K.E.; Ng, C.C.; Abbott, C.A.; Bowling, F.L.; Rajbhandari, S.; Boulton, A.J.M.; Reeves, N.D. A New Mobile Application for Standardizing Diabetic Foot Images. J. Diabetes Sci. Technol. 2018, 12, 169–173. [Google Scholar] [CrossRef] [Green Version]
- Afsaneh, E.; Sharifdini, A.; Ghazzaghi, H.; Ghobadi, M.Z. Recent applications of machine learning and deep learning models in the prediction, diagnosis, and management of diabetes: A comprehensive review. Diabetol. Metab. Syndr. 2022, 14, 196. [Google Scholar] [CrossRef]
- Fregoso-Aparicio, L.; Noguez, J.; Montesinos, L.; García-García, J.A. Machine learning and deep learning predictive models for type 2 diabetes: A systematic review. Diabetol. Metab. Syndr. 2021, 13, 148. [Google Scholar] [CrossRef]
- Abbas, H.T.; Alic, L.; Erraguntla, M.; Ji, J.X.; Abdul-Ghani, M.; Abbasi, Q.H.; Qaraqe, M.K. Predicting long-term type 2 diabetes with support vector machine using oral glucose tolerance test. PLoS ONE 2019, 14, e0219636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ismail, L.; Materwala, H.; Al Kaabi, J. Association of risk factors with type 2 diabetes: A systematic review. Comput. Struct. Biotechnol. J. 2021, 19, 1759–1785. [Google Scholar] [CrossRef]
- National Health and Nutrition Examination Survey. National Center for Health Statistics, 1999–2018. Available online: https://www.cdc.gov/nchs/nhanes/index.htm (accessed on 15 February 2023).
- Johnson, A.; Pollard, T.; Mark, R. MIMIC-III Clinical Database (version 1.4). PhysioNet 2016. [Google Scholar] [CrossRef]
- Johnson, A.; Bulgarelli, L.; Pollard, T.; Horng, S.; Celi, L.A.; Mark, R. MIMIC-IV (version 2.1). PhysioNet 2022. [Google Scholar] [CrossRef]
- De, S.; Mukherjee, A.; Ullah, E. Convergence guarantees for RMSProp and Adam in non-convex optimization and and empirical comparison to Nesterov acceleration. arXiv 2018, arXiv:1807.06766. [Google Scholar]
- Hinton, G. Coursera Neural Networks for Machine Learning Lecture 6, 2018. Available online: https://www.coursera.org/learn/neural-networks-deep-learning (accessed on 15 February 2023).
- Bishop, C. Neural Networks for Pattern Recognition; Oxford University Press: Oxford, UK, 1995; ISBN 9780198538646. [Google Scholar]
- Bitzur, R.; Cohen, H.; Kamari, Y.; Shaish, A.; Harats, D. Triglycerides and HDL cholesterol: Stars or second leads in diabetes? Diabetes Care 2009, 32 (Suppl. S2), S373–S377. [Google Scholar] [CrossRef] [Green Version]
- Muhammad, I.F.; Bao, X.; Nilsson, P.M.; Zaigham, S. Triglyceride-glucose (TyG) index is a predictor of arterial stiffness, incidence of diabetes, cardiovascular disease, and all-cause and cardiovascular mortality: A longitudinal two-cohort analysis. Front. Cardiovasc. Med. 2023, 9, 1035105. [Google Scholar] [CrossRef]
- Aikens, R.C.; Zhao, W.; Saleheen, D.; Reilly, M.P.; Epstein, S.E.; Tikkanen, E.; Salomaa, V.; Voight, B.F. Systolic Blood Pressure and Risk of Type 2 Diabetes: A Mendelian Randomization Study. Diabetes 2017, 66, 543–550. [Google Scholar] [CrossRef] [Green Version]
- Malone, J.I.; Hansen, B.C. Does obesity cause type 2 diabetes mellitus (T2DM)? Or is it the opposite? Pediatr. Diabetes 2019, 20, 5–9. [Google Scholar] [CrossRef] [Green Version]
- Gray, N.; Picone, G.; Sloan, F.; Yashkin, A. Relation between BMI and diabetes mellitus and its complications among US older adults. South Med. J. 2015, 108, 29–36. [Google Scholar] [CrossRef] [Green Version]
- Kautzky-Willer, A.; Harreiter, J.; Pacini, G. Sex and Gender Differences in Risk, Pathophysiology and Complications of Type 2 Diabetes Mellitus. Endocr. Rev. 2016, 37, 278–316. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Yang, P.; Yan, Z.; Liu, Z.; Ma, Q.; Zhang, Z.; Wang, Y.; Su, Y. The Relationship between Erythrocytes and Diabetes Mellitus. J. Diabetes Res. 2021, 2021, 6656062. [Google Scholar] [CrossRef] [PubMed]
- Hornik, K.; Stinchcombe, M.; White, H. Multilayer feedforward networks are universal approximators. Neural Netw. 1989, 2, 359–366. [Google Scholar] [CrossRef]
- Elveren, E.; Yumuşak, N. Tuberculosis disease diagnosis using artificial neural network trained with genetic algorithm. J. Med. Syst. 2011, 35, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Ercal, F.; Chawla, A.; Stoecker, W.V.; Lee, H.C.; Moss, R.H. Neural network diagnosis of malignant melanoma from color images. IEEE Trans. Biomed. Eng. 1994, 41, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Cangelosi, D.; Pelassa, S.; Morini, M.; Conte, M.; Bosco, M.C.; Eva, A.; Sementa, A.R.; Varesio, L. Artificial neural network classifier predicts neuroblastoma patients’ outcome. BMC Bioinform. 2016, 17 (Suppl. S12), 347. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Raoof, M.; Montgomery, S.; Ottosson, J.; Näslund, I. Predicting Long-Term Health-Related Quality of Life after Bariatric Surgery Using a Conventional Neural Network: A Study Based on the Scandinavian Obesity Surgery Registry. J. Clin. Med. 2019, 8, 2149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Courcoulas, A.P.; Christian, N.J.; O’Rourke, R.W.; Dakin, G.; Patchen Dellinger, E.; Flum, D.R.; Melissa Kalarchian, P.D.; Mitchell, J.E.; Patterson, E.; Pomp, A.; et al. Preoperative factors and 3-year weight change in the Longitudinal Assessment of Bariatric Surgery (LABS) consortium. Surg. Obes. Relat. Dis. 2015, 11, 1109–1118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatoum, I.J.; Blackstone, R.; Hunter, T.D.; Francis, D.M.; Steinbuch, M.; Harris, J.L.; Kaplan, L.M. Clinical Factors Associated With Remission of Obesity-Related Comorbidities After Bariatric Surgery. JAMA Surg. 2016, 151, 130–137. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Kiik, M.; Peek, N.; Curcin, V.; Marshall, I.J.; Rudd, A.G.; Wang, Y.; Douiri, A.; Wolfe, C.D.; Bray, B. A systematic review of machine learning models for predicting outcomes of stroke with structured data. PLoS ONE 2020, 15, e0234722. [Google Scholar] [CrossRef]
- Dormann, C.F. Calibration of probability predictions from machine-learning and statistical models. Glob. Ecol Biogeogr. 2020, 29, 760–765. [Google Scholar] [CrossRef]
- Van Calster, B.; Vickers, A.J. Calibration of Risk Prediction Models: Impact on Decision-Analytic Performance. Med. Decis. Mak. 2015, 35, 162–169. [Google Scholar] [CrossRef]





| Feature | Count | Mean | Standard Deviation | Minimum Value | Maximum Value |
|---|---|---|---|---|---|
| Gender/Sex a | 52,640 | 1.512 | 0.499 | 1.0 | 2.0 |
| Age (years) | 52,640 | 43.764 | 24 336 | 12.0 | 300.0 |
| Diabetes b | 52,640 | 0.130 | 0.337 | 0.0 | 1.0 |
| HDL-cholesterol (mg/dL) | 52,640 | 52.112 | 15.535 | 3.0 | 226.0 |
| Glucose (mg/dL) | 52,640 | 103.255 | 36.607 | 21.0 | 683.0 |
| Systolic: Blood pres (mm/Hg) | 52,640 | 121.440 | 18.820 | 51.0 | 270.0 |
| Diastolic: Blood pres (mm/Hg) | 52,640 | 68.041 | 12.938 | 21.9 | 676.1 |
| Triglycerides (mg/dL) | 52,640 | 139.093 | 122.210 | 9.0 | 6057.0 |
| Weight (kg) | 52,640 | 78.153 | 21.506 | 25.1 | 371.0 |
| Body Mass Index (kg/m2) | 52,640 | 33.061 | 948.424 | 3.24 | 215.7 |
| Feature | Count | Mean | Standard Deviation | Minimum Value | Maximum Value |
|---|---|---|---|---|---|
| Gender/Sex a | 13,687 | 1.543 | 0.498 | 1.0 | 2.0 |
| Age (years) | 13,687 | 51.947 | 21.179 | 12.0 | 99.0 |
| Diabetes b | 13,687 | 0.498 | 0.500 | 0.0 | 1.0 |
| HDL-cholesterol (mg/dL) | 13,687 | 49.914 | 15.607 | 3.0 | 158.0 |
| Glucose (mg/dL) | 13,687 | 124.347 | 56.437 | 21.0 | 649.0 |
| Systolic: Blood pres (mm/Hg) | 13,687 | 124.506 | 19.873 | 51.0 | 242.0 |
| Diastolic: Blood pres (mm/Hg) | 13,687 | 67.498 | 12.430 | 21.9 | 202.3 |
| Triglycerides (mg/dL) | 13,687 | 152.276 | 107.965 | 12.0 | 896.0 |
| Weight (kg) | 13,687 | 82.788 | 22.865 | 27.8 | 273.0 |
| Body Mass Index (kg/m2) | 13,687 | 29.456 | 7.313 | 3.2 | 97.4 |
| Hidden Nodes | Accuracy |
|---|---|
| 12 | 0.838 |
| 13 | 0.838 |
| 14 | 0.837 |
| 15 | 0.837 |
| 11 | 0.837 |
| 10 | 0.836 |
| 7 | 0.835 |
| 5 | 0.835 |
| 9 | 0.835 |
| 6 | 0.834 |
| Optimizer | Mean (Accuracy) | Standard Deviation (Accuracy) |
|---|---|---|
| ADAM | 0.855 | 0.008 |
| SGD | 0.853 | 0.009 |
| RMSPROP | 0.852 | 0.009 |
| LM | 0.835 | 0.049 |
| Model | Accuracy |
|---|---|
| SGD | 0.862 |
| RMSPROP | 0.861 |
| ADAM | 0.858 |
| LM | 0.840 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agliata, A.; Giordano, D.; Bardozzo, F.; Bottiglieri, S.; Facchiano, A.; Tagliaferri, R. Machine Learning as a Support for the Diagnosis of Type 2 Diabetes. Int. J. Mol. Sci. 2023, 24, 6775. https://doi.org/10.3390/ijms24076775
Agliata A, Giordano D, Bardozzo F, Bottiglieri S, Facchiano A, Tagliaferri R. Machine Learning as a Support for the Diagnosis of Type 2 Diabetes. International Journal of Molecular Sciences. 2023; 24(7):6775. https://doi.org/10.3390/ijms24076775
Chicago/Turabian StyleAgliata, Antonio, Deborah Giordano, Francesco Bardozzo, Salvatore Bottiglieri, Angelo Facchiano, and Roberto Tagliaferri. 2023. "Machine Learning as a Support for the Diagnosis of Type 2 Diabetes" International Journal of Molecular Sciences 24, no. 7: 6775. https://doi.org/10.3390/ijms24076775
APA StyleAgliata, A., Giordano, D., Bardozzo, F., Bottiglieri, S., Facchiano, A., & Tagliaferri, R. (2023). Machine Learning as a Support for the Diagnosis of Type 2 Diabetes. International Journal of Molecular Sciences, 24(7), 6775. https://doi.org/10.3390/ijms24076775