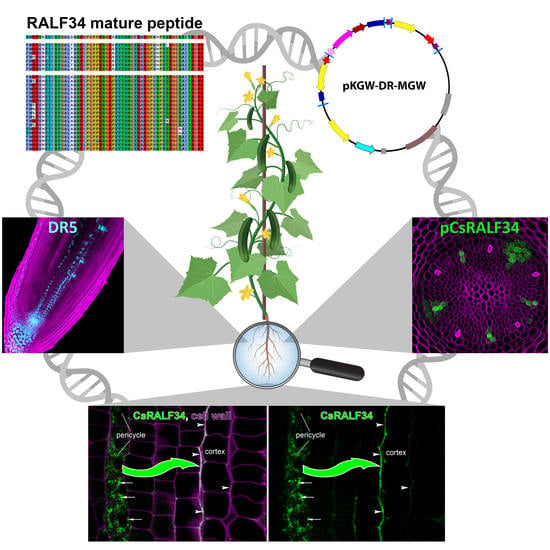
Lateral Root Initiation in Cucumber (Cucumis sativus): What Does the Expression Pattern of Rapid Alkalinization Factor 34 (RALF34) Tell Us?
Abstract
:1. Introduction
2. Results
2.1. Identification of the Ortholog(s) of Arabidopsis Small Signaling Peptide RALF34 and Its Putative Receptor THESEUS1 in Cucumis sativus by Phylogenetic Analysis
2.2. CsRALF34 Is Expressed Ubiquitously in Cucumber; Its Expression Is Not Affected by Auxin but Repressed by Ethylene
2.3. The First Cellular Events in Lateral Root Initiation in Cucumis sativus Roots
2.4. Localization of RALF34 Expression in the Meristem of Cucumis sativus Roots
2.5. CsRALF34 Peptide Accumulated in the Apoplast and Along the Cell Walls of Root Cortex Cells
2.6. The Expression of CsTHE1 Takes Place in All Cells of Cucumis sativus Root Tips
3. Discussion
4. Materials and Methods
4.1. Plant Material and Bacterial Strains
4.2. Phylogeny and Bioinformatics
4.3. Molecular Cloning, Plasmid Construction and Plant Transformation
4.4. Plant Organs Collection
4.5. Treatments with Exogenous Auxin, Precursor of Ethylene and Inhibitor of Ethylene Biosynthesis
4.6. RT-qPCR Assays
4.7. Fluorescence Protein Reporter Assays and Microscopy
4.8. Statistical Analyses
5. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Delay, C.; Imin, N.; Djordjevic, M.A. Regulation of Arabidopsis root development by small signaling peptides. Front. Plant Sci. 2013, 4, 352. [Google Scholar] [CrossRef] [PubMed]
- Jourquin, J.; Fukaki, H.; Beeckman, T. Peptide-receptor signaling controls lateral root development. Plant Physiol. 2020, 182, 1645–1656. [Google Scholar] [CrossRef] [PubMed]
- Gancheva, M.S.; Malovichko, Y.V.; Poliushkevich, L.O.; Dodueva, I.E.; Lutova, L.A. Plant peptide hormones. Russ. J. Plant. Physiol. 2019, 66, 171–189. [Google Scholar] [CrossRef]
- Blackburn, M.R.; Haruta, M.; Moura, D.S. Twenty years of progress in physiological and biochemical investigation of RALF peptides. Plant Physiol. 2020, 182, 1657–1666. [Google Scholar] [CrossRef]
- Campbell, L.; Turner, S.R. A comprehensive analysis of RALF proteins in green plants suggests there are two distinct functional groups. Front. Plant Sci. 2017, 8, 37. [Google Scholar] [CrossRef]
- Atkinson, N.J.; Lilley, C.J.; Urwin, P.E. Identification of genes involved in the response of Arabidopsis to simultaneous biotic and abiotic stresses. Plant Physiol. 2013, 162, 2028–2041. [Google Scholar] [CrossRef]
- Baez, L.A.; Tichá, T.; Hamann, T. Cell wall integrity regulation across plant species. Plant Mol. Biol. 2022, 109, 483–504. [Google Scholar] [CrossRef]
- Murphy, E.; Vu, L.D.; Van den Broeck, L.; Lin, Z.; Ramakrishna, P.; van de Cotte, B.; Gaudinier, A.; Goh, T.; Slane, D.; Beeckman, T.; et al. RALFL34 regulates formative cell divisions in Arabidopsis pericycle during lateral root initiation. J. Exp. Bot. 2016, 67, 4863–4875. [Google Scholar] [CrossRef]
- Bergonci, T.; Ribeiro, B.; Ceciliato, P.H.O.; Guerrero-Abad, J.C.; Silva-Filho, M.C.; Moura, D.S. Arabidopsis thaliana RALF1 opposes brassinosteroid effects on root cell elongation and lateral root formation. J. Exp. Bot. 2014, 65, 2219–2230. [Google Scholar] [CrossRef]
- Gjetting, S.K.; Mahmood, K.; Shabala, L.; Kristensen, A.; Shabala, S.; Palmgren, M.; Fuglsang, A.T. Evidence for multiple receptors mediating RALF-triggered Ca2+ signaling and proton pump inhibition. Plant J. 2020, 104, 433–446. [Google Scholar] [CrossRef]
- Morato do Canto, A.; Ceciliato, P.H.O.; Ribeiro, B.; Ortiz Morea, F.A.; Franco Garcia, A.A.; Silva-Filho, M.C.; Moura, D.S. Biological activity of nine recombinant AtRALF peptides: Implications for their perception and function in Arabidopsis. Plant Physiol. Biochem. 2014, 75, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Abarca, A.; Franck, C.M.; Zipfel, C. Family-wide evaluation of RAPID ALKALINIZATION FACTOR peptides. Plant Physiol. 2021, 187, 996–1010. [Google Scholar] [CrossRef] [PubMed]
- Nikonorova, N.; Murphy, E.; De Lima, C.F.F.; Zhu, S.; Van de Cotte, B.; Vu, L.D.; Balcerowicz, D.; Li, L.; Kong, X.; De Rop, G.; et al. The Arabidopsis root tip (phospho)proteomes at growth-promoting versus growth-repressing conditions reveal novel root growth regulators. Cells 2021, 10, 1665. [Google Scholar] [CrossRef] [PubMed]
- Thieme, C.J.; Rojas-Triana, M.; Stecyk, E.; Schudoma, C.; Zhang, W.; Yang, L.; Miñambres, M.; Walther, D.; Schulze, W.X.; Paz-Ares, J.; et al. Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat. Plants 2015, 1, 15025. [Google Scholar] [CrossRef]
- Bedinger, P.A.; Pearce, G.; Covey, P.A. RALFs: Peptide regulators of plant growth. Plant Signal. Behav. 2010, 5, 1342–1346. [Google Scholar] [CrossRef]
- Escobar, N.M.; Haupt, S.; Thow, G.; Boevink, P.; Chapman, S.; Oparka, K. High-throughput viral expression of cDNA-green fluorescent protein fusions reveals novel subcellular addresses and identifies unique proteins that interact with plasmodesmata. Plant Cell 2003, 15, 1507–1523. [Google Scholar] [CrossRef]
- Chevalier, E.; Loubert-Hudon, A.; Matton, D.P. ScRALF3, a secreted RALF-like peptide involved in cell-cell communication between the sporophyte and the female gametophyte in a solanaceous species. Plant J. 2013, 73, 1019–1033. [Google Scholar] [CrossRef]
- Haruta, M.; Monshausen, G.; Gilroy, S.; Sussman, M.R. A cytoplasmic Ca2+ functional assay for identifying and purifying endogenous cell signaling peptides in Arabidopsis seedlings: Identification of AtRALF1 peptide. Biochemistry 2008, 47, 6311–6321. [Google Scholar] [CrossRef]
- Pearce, G.; Moura, D.S.; Stratmann, J.; Ryan, C.A. RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc. Natl. Acad. Sci. USA 2001, 98, 12843–12847. [Google Scholar] [CrossRef]
- Hirakawa, Y. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. USA 2008, 105, 15208–15213. [Google Scholar] [CrossRef]
- Gonneau, M.; Desprez, T.; Martin, M.; Doblas, V.G.; Bacete, L.; Miart, F.; Sormani, R.; Hématy, K.; Renou, J.; Landrein, B.; et al. Receptor kinase THESEUS1 is a Rapid Alkalinization Factor 34 receptor in Arabidopsis. Curr. Biol. 2018, 28, 2452–2458. [Google Scholar] [CrossRef] [PubMed]
- Merz, D.; Richter, J.; Gonneau, M.; Sanchez-Rodriguez, C.; Eder, T.; Sormani, R.; Martin, M.; Hématy, K.; Höfte, H.; Hauser, M.-T. T-DNA alleles of the receptor kinase THESEUS1 with opposing effects on cell wall integrity signaling. J. Exp. Bot. 2017, 68, 4583–4593. [Google Scholar] [CrossRef] [PubMed]
- Hématy, K.; Sado, P.-E.; Van Tuinen, A.; Rochange, S.; Desnos, T.; Balzergue, S.; Pelletier, S.; Renou, J.-P.; Höfte, H. A Receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr. Biol. 2007, 17, 922–931. [Google Scholar] [CrossRef] [PubMed]
- Nissen, K.S.; Willats, W.G.T.; Malinovsky, F.G. Understanding CrRLK1L function: Cell walls and growth control. Trends Plant Sci. 2016, 21, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Hematy, K.; Hofte, H. Novel receptor kinases involved in growth regulation. Curr. Opin. Plant Biol. 2008, 11, 321–328. [Google Scholar] [CrossRef]
- Boisson-Dernier, A.; Kessler, S.A.; Grossniklaus, U. The walls have ears: The role of plant CrRLK1Ls in sensing and transducing extracellular signals. J. Exp. Bot. 2011, 62, 1581–1591. [Google Scholar] [CrossRef]
- Haruta, M.; Sabat, G.; Stecker, K.; Minkoff, B.B.; Sussman, M.R. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 2014, 343, 408–411. [Google Scholar] [CrossRef]
- Zhu, S.; Martínez Pacheco, J.; Estevez, J.M.; Yu, F. Autocrine regulation of root hair size by the RALF-FERONIA-RSL4 signaling pathway. New Phytol. 2020, 227, 45–49. [Google Scholar] [CrossRef]
- Torres-Martínez, H.H.; Napsucialy-Mendivil, S.; Dubrovsky, J.G. Cellular and molecular bases of lateral root initiation and morphogenesis. Curr. Opin. Plant Biol. 2022, 65, 102115. [Google Scholar] [CrossRef]
- Demchenko, K.N.; Demchenko, N.P. Changes of Root Structure in Connection with the Development of Lateral Root Primordia in Wheat and Pumpkins. In Recent Advances of Plant Root Structure and Function; Developments in Plant and Soil, Sciences; Gašparíková, O., Čiamporová, M., Mistrík, I., Baluška, F., Eds.; Springer: Dordrecht, The Netherlands, 2001; Volume 90, pp. 39–47. [Google Scholar]
- Ilina, E.L.; Kiryushkin, A.S.; Semenova, V.A.; Demchenko, N.P.; Pawlowski, K.; Demchenko, K.N. Lateral root initiation and formation within the parental root meristem of Cucurbita pepo: Is auxin a key player? Ann. Bot. 2018, 122, 873–888. [Google Scholar] [CrossRef]
- Kiryushkin, A.S.; Ilina, E.L.; Puchkova, V.A.; Guseva, E.D.; Pawlowski, K.; Demchenko, K.N. Lateral root initiation in the parental root meristem of cucurbits: Old players in a new position. Front. Plant Sci. 2019, 10, 365. [Google Scholar] [CrossRef]
- Dubrovsky, J.G. The origin of tissues of the embryo lateral root in the cucumber. Interactions between tissues and positional control in its development. Ontogenez Russ. J. Dev. Biol. 1986, 17, 176–189. [Google Scholar]
- De Rybel, B.; Vassileva, V.; Parizot, B.; Demeulenaere, M.; Grunewald, W.; Audenaert, D.; Van Campenhout, J.; Overvoorde, P.; Jansen, L.; Vanneste, S.; et al. A novel Aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr. Biol. 2010, 20, 1697–1706. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Liu, X.; Yan, S.; Liu, B.; Zhong, Y.; Song, W.; Chen, J.; Wang, Z.; Che, G.; Liu, L.; et al. Pollen tube emergence is mediated by ovary-expressed ALCATRAZ in cucumber. Nat. Commun. 2023, 14, 258. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Schiefelbein, J. Conserved gene expression programs in developing roots from diverse plants. Plant Cell 2015, 27, 2119–2132. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, Z.; Yan, P.; Huang, S.; Fei, Z.; Lin, K. RNA-Seq improves annotation of protein-coding genes in the cucumber genome. BMC Genom. 2011, 12, 540. [Google Scholar] [CrossRef]
- Wei, G.; Tian, P.; Zhang, F.; Qin, H.; Miao, H.; Chen, Q.; Hu, Z.; Cao, L.; Wang, M.; Gu, X.; et al. Integrative analyses of nontargeted volatile profiling and transcriptome data provide molecular insight into VOC diversity in cucumber plants (Cucumis sativus). Plant Physiol. 2016, 172, 603–618. [Google Scholar] [CrossRef]
- Richter, J.; Watson, J.M.; Stasnik, P.; Borowska, M.; Neuhold, J.; Berger, M.; Stolt-Bergner, P.; Schoft, V.; Hauser, M.-T. Multiplex mutagenesis of four clustered CrRLK1L with CRISPR/Cas9 exposes their growth regulatory roles in response to metal ions. Sci. Rep. 2018, 8, 12182. [Google Scholar] [CrossRef]
- Omelyanchuk, N.A.; Wiebe, D.S.; Novikova, D.D.; Levitsky, V.G.; Klimova, N.; Gorelova, V.; Weinholdt, C.; Vasiliev, G.V.; Zemlyanskaya, E.V.; Kolchanov, N.A.; et al. Auxin regulates functional gene groups in a fold-change-specific manner in Arabidopsis thaliana roots. Sci. Rep. 2017, 7, 2489. [Google Scholar] [CrossRef]
- Stigliani, A.; Martin-Arevalillo, R.; Lucas, J.; Bessy, A.; Vinos-Poyo, T.; Mironova, V.; Vernoux, T.; Dumas, R.; Parcy, F. Capturing auxin response factors syntax using DNA binding models. Mol. Plant 2018, 12, 822–832. [Google Scholar] [CrossRef]
- Satoh, S.; Esashi, Y. α-Aminoisobutyric acid: A probable competitive inhibitor of conversion of 1-aminocyclopropane-1-carboxylic acid to ethylene. Plant Cell Physiol. 1980, 21, 939–949. [Google Scholar] [CrossRef]
- Adams, D.O.; Yang, S.F. Ethylene biosynthesis: Identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc. Natl. Acad. Sci. USA 1979, 76, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Torres-Martínez, H.H.; Hernández-Herrera, P.; Corkidi, G.; Dubrovsky, J.G. From one cell to many: Morphogenetic field of lateral root founder cells in Arabidopsis thaliana is built by gradual recruitment. Proc. Natl. Acad. Sci. USA 2020, 117, 20943–20949. [Google Scholar] [CrossRef] [PubMed]
- Xuan, W.; De Gernier, H.; Beeckman, T. The dynamic nature and regulation of the root clock. Development 2020, 147, dev181446. [Google Scholar] [CrossRef] [PubMed]
- De Smet, I.; Tetsumura, T.; De Rybel, B.; Frey, N.F.d.; Laplaze, L.; Casimiro, I.; Swarup, R.; Naudts, M.; Vanneste, S.; Audenaert, D.; et al. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 2007, 134, 681–690. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Risueno, M.A.; Van Norman, J.M.; Moreno, A.; Zhang, J.; Ahnert, S.E.; Benfey, P.N. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 2010, 329, 1306–1311. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, T.; Yalamanchili, K.; de Gernier, H.; Santos Teixeira, J.; Beeckman, T.; Scheres, B.; Willemsen, V.; ten Tusscher, K. A reflux-and-growth mechanism explains oscillatory patterning of lateral root branching sites. Dev. Cell 2021, 56, 2176–2191. [Google Scholar] [CrossRef]
- Laskowski, M.; ten Tusscher, K.H. Periodic Lateral Root Priming: What Makes It Tick? Plant Cell 2017, 29, 432–444. [Google Scholar] [CrossRef]
- Liao, C.-Y.; Smet, W.; Brunoud, G.; Yoshida, S.; Vernoux, T.; Weijers, D. Reporters for sensitive and quantitative measurement of auxin response. Nat. Methods 2015, 12, 207–210. [Google Scholar] [CrossRef]
- Xuan, W.; Audenaert, D.; Parizot, B.; Möller, B.K.; Njo, M.F.; De Rybel, B.; De Rop, G.; Van Isterdael, G.; Mähönen, A.P.; Vanneste, S.; et al. Root cap-derived auxin pre-patterns the longitudinal axis of the Arabidopsis root. Curr. Biol. 2015, 25, 1381–1388. [Google Scholar] [CrossRef]
- Xuan, W.; Band, L.R.; Kumpf, R.P.; Van Damme, D.; Parizot, B.; De Rop, G.; Opdenacker, D.; Möller, B.K.; Skorzinski, N.; Njo, M.F.; et al. Cyclic programmed cell death stimulates hormone signaling and root development in Arabidopsis. Science 2016, 351, 384–387. [Google Scholar] [CrossRef] [PubMed]
- Xuan, W.; Opdenacker, D.; Vanneste, S.; Beeckman, T. Long-Term In Vivo Imaging of Luciferase-Based Reporter Gene Expression in Arabidopsis Roots. In Root Development: Methods and Protocols; Ristova, D., Barbez, E., Eds.; Springer: New York, NY, USA, 2018; pp. 177–190. [Google Scholar] [CrossRef]
- Motte, H.; Vanneste, S.; Beeckman, T. Molecular and environmental regulation of root development. Annu. Rev. Plant Biol. 2019, 70, 465–488. [Google Scholar] [CrossRef] [PubMed]
- Dubrovsky, J.G.; Rost, T.L.; Colon-Carmona, A.; Doerner, P. Early primordium morphogenesis during lateral root initiation in Arabidopsis thaliana. Planta 2001, 214, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Dubrovsky, J.G.; Sauer, M.; Napsucialy-Mendivil, S.; Ivanchenko, M.G.; Friml, J.; Shishkova, S.; Celenza, J.; Benkova, E. Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc. Natl. Acad. Sci. USA 2008, 105, 8790–8794. [Google Scholar] [CrossRef] [PubMed]
- Casero, P.J.; Casimiro, I.; Rodriguez-Gallardo, L.; Martin-Partigo, G.; Lloret, P.G. Lateral root initiation by asymmetrical transverse divisions of pericycle cells in adventitious roots of Allium cepa. Protoplasma 1993, 176, 138–144. [Google Scholar] [CrossRef]
- Casero, P.J.; Casimiro, I.; Lloret, P.G. Lateral root initiation by asymmetrical transverse divisions of pericycle cells in four plant species—Raphanus sativus, Helianthus annuus, Zea mays, and Daucus carota. Protoplasma 1995, 188, 49–58. [Google Scholar] [CrossRef]
- Li, C.; Yeh, F.-L.; Cheung, A.Y.; Duan, Q.; Kita, D.; Liu, M.-C.; Maman, J.; Luu, E.J.; Wu, B.W.; Gates, L.; et al. Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in Arabidopsis. eLife 2015, 4, e06587. [Google Scholar] [CrossRef]
- White, F.F.; Taylor, B.H.; Huffman, G.A.; Gordon, M.P.; Nester, E.W. Molecular and genetic analysis of the transferred DNA regions of the root-inducing plasmid of Agrobacterium rhizogenes. J. Bacteriol. 1985, 164, 33–44. [Google Scholar] [CrossRef]
- Owens, L.D.; Cress, D.E. Genotypic variability of soybean response to Agrobacterium strains harboring the Ti or Ri plasmids. Plant Physiol. 1985, 77, 87–94. [Google Scholar] [CrossRef]
- Berardini, T.Z.; Reiser, L.; Li, D.; Mezheritsky, Y.; Muller, R.; Strait, E.; Huala, E. The arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 2015, 53, 474–485. [Google Scholar] [CrossRef]
- Zheng, Y.; Wu, S.; Bai, Y.; Sun, H.; Jiao, C.; Guo, S.; Zhao, K.; Blanca, J.; Zhang, Z.; Huang, S.; et al. Cucurbit Genomics Database (CuGenDB): A central portal for comparative and functional genomics of cucurbit crops. Nucleic Acids Res. 2019, 47, D1128–D1136. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Koo, D.-H.; Li, Y.; Zhang, X.; Luan, F.; Havey, M.J.; Jiang, J.; Weng, Y. Chromosome rearrangements during domestication of cucumber as revealed by high-density genetic mapping and draft genome assembly. Plant J. 2012, 71, 895–906. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: Approximate methods. J. Mol. Evol. 1994, 39, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Shoemaker, J.S.; Fitch, W.M. Evidence from nuclear sequences that invariable sites should be considered when sequence divergence is calculated. Mol. Biol. Evol. 1989, 6, 270–289. [Google Scholar] [CrossRef] [PubMed]
- Whelan, S.; Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef]
- Schwarz, G. Estimating the dimension of a model. Ann. Stat. 1978, 6, 461–464. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Gascuel, O. BIONJ: An improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 1997, 14, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Ilina, E.L.; Kiryushkin, A.S.; Demchenko, K.N. Features of fluorescent protein application to study the root system development of cucurbits (Cucurbitaceae). Agric. Biol. Sel’skokhozyaistvennaya Biol. 2020, 55, 1040–1055. [Google Scholar] [CrossRef]
- Limpens, E.; Ramos, J.; Franken, C.; Raz, V.; Compaan, B.; Franssen, H.; Bisseling, T.; Geurts, R. RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J. Exp. Bot. 2004, 55, 983–992. [Google Scholar] [CrossRef] [PubMed]
- Ulmasov, T.; Murfett, J.; Hagen, G.; Guilfoyle, T.J. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 1997, 9, 1963–1971. [Google Scholar] [CrossRef]
- Hornung, E.; Krueger, C.; Pernstich, C.; Gipmans, M.; Porzel, A.; Feussner, I. Production of (10E,12Z)-conjugated linoleic acid in yeast and tobacco seeds. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2005, 1738, 105–114. [Google Scholar] [CrossRef]
- Shaner, N.C.; Lambert, G.G.; Chammas, A.; Ni, Y.; Cranfill, P.J.; Baird, M.A.; Sell, B.R.; Allen, J.R.; Day, R.N.; Israelsson, M.; et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 2013, 10, 407–409. [Google Scholar] [CrossRef]
- Ilina, E.L.; Logachov, A.A.; Laplaze, L.; Demchenko, N.P.; Pawlowski, K.; Demchenko, K.N. Composite Cucurbita pepo plants with transgenic roots as a tool to study root development. Ann. Bot. 2012, 110, 479–489. [Google Scholar] [CrossRef]
- Hoagland, D.R.; Arnon, D.T. The Water-Culture Method for Growing Plants without Soil. In California Agriculture Experiment Station; Circular 347; University of California: Berkeley, CA, USA, 1938; pp. 1–39. [Google Scholar]
- Paponov, I.A.; Paponov, M.; Teale, W.; Menges, M.; Chakrabortee, S.; Murray, J.A.H.; Palme, K. Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol. Plant 2008, 1, 321–337. [Google Scholar] [CrossRef]
- Lemaire, L.; Deleu, C.; Le Deunff, E. Modulation of ethylene biosynthesis by ACC and AIB reveals a structural and functional relationship between the K15NO3 uptake rate and root absorbing surfaces. J. Exp. Bot. 2013, 64, 2725–2737. [Google Scholar] [CrossRef] [PubMed]
- Markakis, M.N.; De Cnodder, T.; Lewandowski, M.; Simon, D.; Boron, A.; Balcerowicz, D.; Doubbo, T.; Taconnat, L.; Renou, J.-P.; Höfte, H.; et al. Identification of genes involved in the ACC-mediated control of root cell elongation in Arabidopsis thaliana. BMC Plant Biol. 2012, 12, 208. [Google Scholar] [CrossRef] [PubMed]
- Harkey, A.F.; Watkins, J.M.; Olex, A.L.; DiNapoli, K.T.; Lewis, D.R.; Fetrow, J.S.; Binder, B.M.; Muday, G.K. Identification of transcriptional and receptor networks that control root responses to ethylene. Plant Physiol. 2017, 176, 2095–2118. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Wan, H.; Zhao, Z.; Qian, C.; Sui, Y.; Malik, A.A.; Chen, J. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal. Biochem. 2010, 399, 257–261. [Google Scholar] [CrossRef]
- Lebedeva, M.A.; Tvorogova, V.E.; Vinogradova, A.P.; Gancheva, M.S.; Azarakhsh, M.; Ilina, E.L.; Demchenko, K.N.; Dodueva, I.E.; Lutova, L.A. Initiation of spontaneous tumors in radish (Raphanus sativus): Cellular, molecular and physiological events. J. Plant Physiol. 2015, 173, 97–104. [Google Scholar] [CrossRef]
- Kiryushkin, A.S.; Ilina, E.L.; Guseva, E.D.; Pawlowski, K.; Demchenko, K.N. Hairy CRISPR: Genome editing in plants using hairy root transformation. Plants 2022, 11, 51. [Google Scholar] [CrossRef]
- Musielak, T.J.; Schenkel, L.; Kolb, M.; Henschen, A.; Bayer, M. A simple and versatile cell wall staining protocol to study plant reproduction. Plant Reprod. 2015, 28, 161–169. [Google Scholar] [CrossRef]
- Kurihara, D.; Mizuta, Y.; Sato, Y.; Higashiyama, T. ClearSee: A rapid optical clearing reagent for whole-plant fluorescence imaging. Development 2015, 142, 4168–4179. [Google Scholar] [CrossRef]
- Kraus, B.; Ziegler, M.; Wolff, H. Linear Fluorescence Unmixing in Cell Biological Research. In Modern Research and Educational Topics in Microscopy; Méndez-Vilas, A., Díaz, J., Eds.; Formatex: Badajoz, Spain, 2007; pp. 863–872. [Google Scholar]
- R Core Team. R: A Language and eEnvironment for Statistical Computing. Available online: https://www.R-project.org (accessed on 1 May 2023).
- Shumilina, J.; Kiryushkin, A.S.; Frolova, N.; Mashkina, V.; Ilina, E.L.; Puchkova, V.A.; Danko, K.; Silinskaya, S.; Serebryakov, E.B.; Soboleva, A.; et al. Integrative proteomics and metabolomics analysis reveals the role of small signaling peptide Rapid Alkalinization Factor 34 (RALF34) in cucumber roots. Int. J. Mol. Sci. 2023, 24, 7654. [Google Scholar] [CrossRef] [PubMed]
- Gogolev, Y.V.; Ahmar, S.; Akpinar, B.A.; Budak, H.; Kiryushkin, A.S.; Gorshkov, V.Y.; Hensel, G.; Demchenko, K.N.; Kovalchuk, I.; Mora-Poblete, F.; et al. OMICs, epigenetics, and genome editing techniques for food and nutritional security. Plants 2021, 10, 1423. [Google Scholar] [CrossRef] [PubMed]
- Argyris, J.M.; Ruiz-Herrera, A.; Madriz-Masis, P.; Sanseverino, W.; Morata, J.; Pujol, M.; Ramos-Onsins, S.E.; Garcia-Mas, J. Use of targeted SNP selection for an improved anchoring of the melon (Cucumis melo L.) scaffold genome assembly. BMC Genom. 2015, 16, 4. [Google Scholar] [CrossRef] [PubMed]
- Barrera-Redondo, J.; Ibarra-Laclette, E.; Vázquez-Lobo, A.; Gutiérrez-Guerrero, Y.T.; Sánchez de la Vega, G.; Piñero, D.; Montes-Hernández, S.; Lira-Saade, R.; Eguiarte, L.E. The genome of Cucurbita argyrosperma (Silver-Seed Gourd) reveals faster rates of protein-coding gene and long noncoding rna turnover and neofunctionalization within Cucurbita. Mol. Plant 2019, 12, 506–520. [Google Scholar] [CrossRef]
- Fu, A.; Wang, Q.; Mu, J.; Ma, L.; Wen, C.; Zhao, X.; Gao, L.; Li, J.; Shi, K.; Wang, Y.; et al. Combined genomic, transcriptomic, and metabolomic analyses provide insights into chayote (Sechium edule) evolution and fruit development. Hortic. Res. 2021, 8, 35. [Google Scholar] [CrossRef]
- Griesmann, M.; Chang, Y.; Liu, X.; Song, Y.; Haberer, G.; Crook, M.B.; Billault-Penneteau, B.; Lauressergues, D.; Keller, J.; Imanishi, L.; et al. Phylogenomics reveals multiple losses of nitrogen-fixing root nodule symbiosis. Science 2018, 361, eaat1743. [Google Scholar] [CrossRef]
- Guo, S.; Zhang, J.; Sun, H.; Salse, J.; Lucas, W.J.; Zhang, H.; Zheng, Y.; Mao, L.; Ren, Y.; Wang, Z.; et al. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat. Genet. 2013, 45, 51–58. [Google Scholar] [CrossRef]
- Huang, D.; Ming, R.; Xu, S.; Wang, J.; Yao, S.; Li, L.; Huang, R.; Tan, Y. Chromosome-level genome assembly of Gynostemma pentaphyllum provides insights into gypenoside biosynthesis. DNA Research 2021, 28, 1–9. [Google Scholar] [CrossRef]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef]
- Li, L.; Chen, X.; Fang, D.; Dong, S.; Guo, X.; Li, N.; Campos-Dominguez, L.; Wang, W.; Liu, Y.; Lang, X.; et al. Genomes shed light on the evolution of Begonia, a mega-diverse genus. New Phytol. 2022, 234, 295–310. [Google Scholar] [CrossRef]
- Ling, J.; Xie, X.; Gu, X.; Zhao, J.; Ping, X.; Li, Y.; Yang, Y.; Mao, Z.; Xie, B. High-quality chromosome-level genomes of Cucumis metuliferus and Cucumis melo provide insight into Cucumis genome evolution. Plant J. 2021, 107, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Wang, Q.; Mu, J.; Fu, A.; Wen, C.; Zhao, X.; Gao, L.; Li, J.; Shi, K.; Wang, Y.; et al. The genome and transcriptome analysis of snake gourd provide insights into its evolution and fruit development and ripening. Hortic. Res. 2020, 7, 199. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, H.; Hsiao, M.-C.; Lin, Y.-P.; Toyoda, A.; Taniai, N.; Tarora, K.; Urasaki, N.; Anand, S.S.; Dhillon, N.P.S.; Schafleitner, R.; et al. Long-read bitter gourd (Momordica charantia) genome and the genomic architecture of nonclassic domestication. Proc. Natl. Acad. Sci. USA 2020, 117, 14543–14551. [Google Scholar] [CrossRef] [PubMed]
- Montero-Pau, J.; Blanca, J.; Bombarely, A.; Ziarsolo, P.; Esteras, C.; Martí-Gómez, C.; Ferriol, M.; Gómez, P.; Jamilena, M.; Mueller, L.; et al. De novo assembly of the zucchini genome reveals a whole-genome duplication associated with the origin of the Cucurbita genus. Plant Biotechnol. J. 2018, 16, 1161–1171. [Google Scholar] [CrossRef]
- Pootakham, W.; Sonthirod, C.; Naktang, C.; Nawae, W.; Yoocha, T.; Kongkachana, W.; Sangsrakru, D.; Jomchai, N.; U-thoomporn, S.; Sheedy, J.R.; et al. De novo assemblies of Luffa acutangula and Luffa cylindrica genomes reveal an expansion associated with substantial accumulation of transposable elements. Mol. Ecol. Resour. 2021, 21, 212–225. [Google Scholar] [CrossRef]
- Qin, X.; Zhang, Z.; Lou, Q.; Xia, L.; Li, J.; Li, M.; Zhou, J.; Zhao, X.; Xu, Y.; Li, Q.; et al. Chromosome-scale genome assembly of Cucumis hystrix—A wild species interspecifically cross-compatible with cultivated cucumber. Hortic. Res. 2021, 8, 40. [Google Scholar] [CrossRef]
- Sun, H.; Wu, S.; Zhang, G.; Jiao, C.; Guo, S.; Ren, Y.; Zhang, J.; Zhang, H.; Gong, G.; Jia, Z.; et al. Karyotype stability and unbiased fractionation in the paleo-allotetraploid Cucurbita genomes. Mol. Plant 2017, 10, 1293–1306. [Google Scholar] [CrossRef]
- Wu, S.; Shamimuzzaman, M.; Sun, H.; Salse, J.; Sui, X.; Wilder, A.; Wu, Z.; Levi, A.; Xu, Y.; Ling, K.-S.; et al. The bottle gourd genome provides insights into Cucurbitaceae evolution and facilitates mapping of a Papaya ring-spot virus resistance locus. Plant J. 2017, 92, 963–975. [Google Scholar] [CrossRef]
- Xia, M.; Han, X.; He, H.; Yu, R.; Zhen, G.; Jia, X.; Cheng, B.; Deng, X.W. Improved de novo genome assembly and analysis of the Chinese cucurbit Siraitia grosvenorii, also known as monk fruit or luo-han-guo. GigaScience 2018, 7, 1–9. [Google Scholar] [CrossRef]
- Xie, D.; Xu, Y.; Wang, J.; Liu, W.; Zhou, Q.; Luo, S.; Huang, W.; He, X.; Li, Q.; Peng, Q.; et al. The wax gourd genomes offer insights into the genetic diversity and ancestral cucurbit karyotype. Nat. Commun. 2019, 10, 5158. [Google Scholar] [CrossRef]
- Zhang, T.; Ren, X.; Zhang, Z.; Ming, Y.; Yang, Z.; Hu, J.; Li, S.; Wang, Y.; Sun, S.; Sun, K.; et al. Long-read sequencing and de novo assembly of the Luffa cylindrica (L.) Roem. genome. Mol. Ecol. Resour. 2020, 20, 511–519. [Google Scholar] [CrossRef] [PubMed]









| Name of Vector | Fusion Construct | Vector Purpose |
|---|---|---|
| pKGW-RR-MGW-pCsRALF34::mNeonGreen-H2B | pCsRALF34::mNeonGreen-H2B | CsRALF34 gene expression analysis |
| pKGW-RR-MGW-pCsRALF34::CsRALF34-mNeonGreen | pCsRALF34::CsRALF34-mNeonGreen | CsRALF34 protein distribution analysis, CsRALF34 fused to mNeonGreen without linker |
| pKGW-RR-MGW-pCsRALF34::CsRALF34-linker-mNeonGreen | pCsRALF34::CsRALF34-linker- mNeonGreen | CsRALF34 protein distribution analysis, CsRALF34 fused to mNeonGreen via linker |
| pKGW-DR-MGW | DR5::mRuby-H2B in backbone | auxin response maxima via DR5 reporter with nuclear localization |
| pKGW-DR-MGW- pCsRALF34::CsRALF34-mNeonGreen | pCsRALF34::CsRALF34-mNeonGreen, DR5::mRuby-H2B | CsRALF34 protein distribution analysis (CsRALF34 fused to mNeonGreen without linker) in combination with DR5-reported auxin response maxima |
| pKGW-RR-MGW- pCsTHESEUS1::mNeonGreen-H2B | pCsTHESEUS1::mNeonGreen-H2B | CsTHESEUS1 gene expression analysis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiryushkin, A.S.; Ilina, E.L.; Guseva, E.D.; Pawlowski, K.; Demchenko, K.N. Lateral Root Initiation in Cucumber (Cucumis sativus): What Does the Expression Pattern of Rapid Alkalinization Factor 34 (RALF34) Tell Us? Int. J. Mol. Sci. 2023, 24, 8440. https://doi.org/10.3390/ijms24098440
Kiryushkin AS, Ilina EL, Guseva ED, Pawlowski K, Demchenko KN. Lateral Root Initiation in Cucumber (Cucumis sativus): What Does the Expression Pattern of Rapid Alkalinization Factor 34 (RALF34) Tell Us? International Journal of Molecular Sciences. 2023; 24(9):8440. https://doi.org/10.3390/ijms24098440
Chicago/Turabian StyleKiryushkin, Alexey S., Elena L. Ilina, Elizaveta D. Guseva, Katharina Pawlowski, and Kirill N. Demchenko. 2023. "Lateral Root Initiation in Cucumber (Cucumis sativus): What Does the Expression Pattern of Rapid Alkalinization Factor 34 (RALF34) Tell Us?" International Journal of Molecular Sciences 24, no. 9: 8440. https://doi.org/10.3390/ijms24098440
APA StyleKiryushkin, A. S., Ilina, E. L., Guseva, E. D., Pawlowski, K., & Demchenko, K. N. (2023). Lateral Root Initiation in Cucumber (Cucumis sativus): What Does the Expression Pattern of Rapid Alkalinization Factor 34 (RALF34) Tell Us? International Journal of Molecular Sciences, 24(9), 8440. https://doi.org/10.3390/ijms24098440