Pennogenin 3-O-β-Chacotrioside Attenuates Hypertrophied Lipid Accumulation by Enhancing Mitochondrial Oxidative Capacity
Abstract
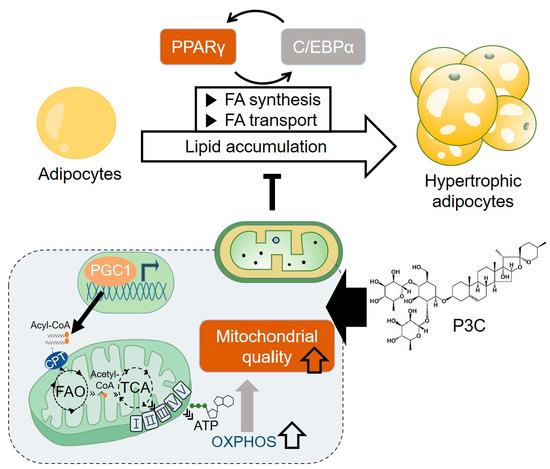
:1. Introduction
2. Results
2.1. P3C Decreased Lipid Accumulation in Hypertrophied 3T3-L1 Adipocytes
2.2. P3C Decreased the Expression of Lipogenesis-Related Genes in Hypertrophied 3T3-L1 Adipocytes
2.3. P3C Increased the Expression of Fatty Acid Oxidation-Related Genes in Hypertrophied 3T3-L1 Adipocytes
2.4. P3C Enhanced the Mitochondrial Oxidative Capacity in Hypertrophied 3T3-L1 Adipocytes
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Cell Culture
4.3. Cell Viability Assay
4.4. Quantification of Lipid Accumulation
4.5. Quantification of Total TG
4.6. Western Blotting Analysis
4.7. Quantitative Real-Time PCR
4.8. Measurement of Oxygen Consumption Rate
4.9. Glycolytic Rate Analysis
4.10. Measurement of Mitochondrial DNA Levels
4.11. Mitochondrial Staining Assay
4.12. Mitochondria Isolation
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lafontan, M. Adipose tissue and adipocyte dysregulation. Diabetes Metab. 2014, 40, 16–28. [Google Scholar] [CrossRef]
- Ipsen, D.H.; Lykkesfeldt, J.; Tveden-Nyborg, P. Molecular mechanisms of hepatic lipid accumulation in non-alcoholic fatty liver disease. Cell. Mol. Life Sci. 2018, 75, 3313–3327. [Google Scholar] [CrossRef]
- Siersbæk, R.; Nielsen, R.; Mandrup, S. PPARγ in adipocyte differentiation and metabolism–novel insights from genome-wide studies. FEBS Lett. 2010, 584, 3242–3249. [Google Scholar] [CrossRef]
- Christodoulides, C.; Vidal-Puig, A. PPARs and adipocyte function. Mol. Cell. Endocrinol. 2010, 318, 61–68. [Google Scholar] [CrossRef]
- Darlington, G.J.; Ross, S.E.; MacDougald, O.A. The role of C/EBP genes in adipocyte differentiation. J. Biol. Chem. 1998, 273, 30057–30060. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.; Pessin, J.E. Adipokines mediate inflammation and insulin resistance. Front. Endocrinol. 2013, 4, 71. [Google Scholar] [CrossRef] [PubMed]
- Liesa, M.; Shirihai, O.S. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 2013, 17, 491–506. [Google Scholar] [CrossRef] [PubMed]
- Melser, S.; Lavie, J.; Bénard, G. Mitochondrial degradation and energy metabolism. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2015, 1853, 2812–2821. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Anand, S.K.; Singh, N.; Dwivedi, U.N.; Kakkar, P. AMP-activated protein kinase: An energy sensor and survival mechanism in the reinstatement of metabolic homeostasis. Exp. Cell Res. 2023, 428, 113614. [Google Scholar] [CrossRef]
- Papa, S.; Martino, P.L.; Capitanio, G.; Gaballo, A.; De Rasmo, D.; Signorile, A.; Petruzzella, V. The oxidative phosphorylation system in mammalian mitochondria. In Advances in Mitochondrial Medicine; Springer: Dordrecht, The Netherlands, 2012; Volume 942, pp. 3–37. [Google Scholar] [CrossRef]
- Nadal-Casellas, A.; Amengual-Cladera, E.; Proenza, A.M.; Lladó, I.; Gianotti, M. Long-term high-fat-diet feeding impairs mitochondrial biogenesis in liver of male and female rats. Cell. Physiol. Biochem. 2010, 26, 291–302. [Google Scholar] [CrossRef]
- Sutherland, L.N.; Capozzi, L.C.; Turchinsky, N.J.; Bell, R.C.; Wright, D.C. Time course of high-fat diet-induced reductions in adipose tissue mitochondrial proteins: Potential mechanisms and the relationship to glucose intolerance. Am. J. Physiol.-Endocrinol. Metab. 2008, 295, E1076–E1083. [Google Scholar] [CrossRef] [PubMed]
- Chiang Morales, M.D.; Chang, C.-Y.; Le, V.L.; Huang, I.-T.; Tsai, I.-L.; Shih, H.-J.; Huang, C.-J. High-fructose/high-fat diet downregulates the hepatic mitochondrial oxidative phosphorylation pathway in mice compared with high-fat diet alone. Cells 2022, 11, 3425. [Google Scholar] [CrossRef] [PubMed]
- Koliaki, C.; Roden, M. Alterations of mitochondrial function and insulin sensitivity in human obesity and diabetes mellitus. Annu. Rev. Nutr. 2016, 36, 337–367. [Google Scholar] [CrossRef] [PubMed]
- Heinonen, S.; Jokinen, R.; Rissanen, A.; Pietiläinen, K.H. White adipose tissue mitochondrial metabolism in health and in obesity. Obes. Rev. 2020, 21, e12958. [Google Scholar] [CrossRef]
- Li, H.; Xu, M.; Lee, J.; He, C.; Xie, Z. Leucine supplementation increases SIRT1 expression and prevents mitochondrial dysfunction and metabolic disorders in high-fat diet-induced obese mice. Am. J. Physiol.-Endocrinol. Metab. 2012, 303, E1234–E1244. [Google Scholar] [CrossRef]
- Hou, C.-Y.; Tain, Y.-L.; Yu, H.-R.; Huang, L.-T. The effects of resveratrol in the treatment of metabolic syndrome. Int. J. Mol. Sci. 2019, 20, 535. [Google Scholar] [CrossRef]
- Yi, X.; Maeda, N. α-Lipoic acid prevents the increase in atherosclerosis induced by diabetes in apolipoprotein E–deficient mice fed high-fat/low-cholesterol diet. Diabetes 2006, 55, 2238–2244. [Google Scholar] [CrossRef]
- Mao, G.; Kraus, G.A.; Kim, I.; Spurlock, M.E.; Bailey, T.B.; Zhang, Q.; Beitz, D.C. A mitochondria-targeted vitamin E derivative decreases hepatic oxidative stress and inhibits fat deposition in mice. J. Nutr. 2010, 140, 1425–1431. [Google Scholar] [CrossRef]
- Hill, J.O.; Wyatt, H.R.; Peters, J.C. Energy balance and obesity. Circulation 2012, 126, 126–132. [Google Scholar] [CrossRef]
- Cusi, K. Role of obesity and lipotoxicity in the development of nonalcoholic steatohepatitis: Pathophysiology and clinical implications. Gastroenterology 2012, 142, 711–725.e716. [Google Scholar] [CrossRef] [PubMed]
- Koliaki, C.; Liatis, S.; Kokkinos, A. Obesity and cardiovascular disease: Revisiting an old relationship. Metabolism 2019, 92, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Apovian, C.M. Obesity: Definition, comorbidities, causes, and burden. Am. J. Manag. Care 2016, 22 (Suppl. 7), s176–s185. [Google Scholar] [PubMed]
- Guglielmi, V.; Bettini, S.; Sbraccia, P.; Busetto, L.; Pellegrini, M.; Yumuk, V.; Colao, A.M.; El Ghoch, M.; Muscogiuri, G. Beyond Weight Loss: Added Benefits Could Guide the Choice of Anti-Obesity Medications. Curr. Obes. Rep. 2023, 12, 127–146. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.T.; Uen, W.C.; Choong, C.Y.; Shi, Y.C.; Lee, B.H.; Tai, C.J.; Tai, C.J. Paris Polyphylla Inhibits Colorectal Cancer Cells via Inducing Autophagy and Enhancing the Efficacy of Chemotherapeutic Drug Doxorubicin. Molecules 2019, 24, 2102. [Google Scholar] [CrossRef]
- Zhang, C.; Jia, X.; Bao, J.; Chen, S.; Wang, K.; Zhang, Y.; Li, P.; Wan, J.-B.; Su, H.; Wang, Y. Polyphyllin VII induces apoptosis in HepG2 cells through ROS-mediated mitochondrial dysfunction and MAPK pathways. BMC Complement. Altern. Med. 2015, 16, 58. [Google Scholar] [CrossRef] [PubMed]
- Kusminski, C.M.; Scherer, P.E. Mitochondrial dysfunction in white adipose tissue. Trends Endocrinol. Metab. 2012, 23, 435–443. [Google Scholar] [CrossRef]
- Lee, J.-E.; Schmidt, H.; Lai, B.; Ge, K. Transcriptional and epigenomic regulation of adipogenesis. Mol. Cell. Biol. 2019, 39, e00601–e00618. [Google Scholar] [CrossRef]
- Tontonoz, P.; Spiegelman, B.M. Fat and beyond: The diverse biology of PPARγ. Annu. Rev. Biochem. 2008, 77, 289–312. [Google Scholar] [CrossRef]
- Boughanem, H.; Cabrera-Mulero, A.; Millán-Gómez, M.; Garrido-Sánchez, L.; Cardona, F.; Tinahones, F.J.; Moreno-Santos, I.; Macías-González, M. Transcriptional Analysis of FOXO1, C/EBP-α and PPAR-γ2 Genes and Their Association with Obesity-Related Insulin Resistance. Genes 2019, 10, 706. [Google Scholar] [CrossRef]
- Lefterova, M.I.; Zhang, Y.; Steger, D.J.; Schupp, M.; Schug, J.; Cristancho, A.; Feng, D.; Zhuo, D.; Stoeckert, C.J.; Liu, X.S. PPARγ and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008, 22, 2941–2952. [Google Scholar] [CrossRef]
- Cheng, C.F.; Ku, H.C.; Lin, H. PGC-1α as a Pivotal Factor in Lipid and Metabolic Regulation. Int. J. Mol. Sci. 2018, 19, 3447. [Google Scholar] [CrossRef]
- Nakamura, M.T.; Yudell, B.E.; Loor, J.J. Regulation of energy metabolism by long-chain fatty acids. Prog. Lipid Res. 2014, 53, 124–144. [Google Scholar] [CrossRef]
- Serra, D.; Mera, P.; Malandrino, M.I.; Mir, J.F.; Herrero, L. Mitochondrial fatty acid oxidation in obesity. Antioxid. Redox Signal. 2013, 19, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Calderon-Dominguez, M.; Sebastian, D.; Fucho, R.; Weber, M.; Mir, J.F.; García-Casarrubios, E.; Obregón, M.J.; Zorzano, A.; Valverde, Á.M.; Serra, D. Carnitine palmitoyltransferase 1 increases lipolysis, UCP1 protein expression and mitochondrial activity in brown adipocytes. PLoS ONE 2016, 11, e0159399. [Google Scholar] [CrossRef] [PubMed]
- Semple, R.; Crowley, V.; Sewter, C.; Laudes, M.; Christodoulides, C.; Considine, R.; Vidal-Puig, A.; O’rahilly, S. Expression of the thermogenic nuclear hormone receptor coactivator PGC-1α is reduced in the adipose tissue of morbidly obese subjects. Int. J. Obes. 2004, 28, 176–179. [Google Scholar] [CrossRef] [PubMed]
- Hammarstedt, A.; Jansson, P.-A.; Wesslau, C.; Yang, X.; Smith, U. Reduced expression of PGC-1 and insulin-signaling molecules in adipose tissue is associated with insulin resistance. Biochem. Biophys. Res. Commun. 2003, 301, 578–582. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Liu, M.; Liu, H.; Xie, J.; Yan, J.; Hou, X.; Liu, J. Zeaxanthin promotes browning by enhancing mitochondrial biogenesis through the PKA pathway in 3T3-L1 adipocytes. Food Funct. 2021, 12, 6283–6293. [Google Scholar] [CrossRef]
- Mu, Q.; Fang, X.; Li, X.; Zhao, D.; Mo, F.; Jiang, G.; Yu, N.; Zhang, Y.; Guo, Y.; Fu, M. Ginsenoside Rb1 promotes browning through regulation of PPARγ in 3T3-L1 adipocytes. Biochem. Biophys. Res. Commun. 2015, 466, 530–535. [Google Scholar] [CrossRef]
- Liu, Z.; Qiao, Q.; Sun, Y.; Chen, Y.; Ren, B.; Liu, X. Sesamol ameliorates diet-induced obesity in C57BL/6J mice and suppresses adipogenesis in 3T3-L1 cells via regulating mitochondria-lipid metabolism. Mol. Nutr. Food Res. 2017, 61, 1600717. [Google Scholar] [CrossRef]
- Bae, I.S.; Kim, S.H. Sinapic Acid Promotes Browning of 3T3-L1 Adipocytes via p38 MAPK/CREB Pathway. Biomed. Res. Int. 2020, 2020, 5753623. [Google Scholar] [CrossRef]
- Ziqubu, K.; Muller, C.J.F.; Dludla, P.V.; Mthembu, S.X.H.; Obonye, N.; Louw, J.; Kappo, A.P.; Silvestri, S.; Orlando, P.; Tiano, L.; et al. Impact of Isoorientin on Metabolic Activity and Lipid Accumulation in Differentiated Adipocytes. Molecules 2020, 25, 1773. [Google Scholar] [CrossRef]
- Pan, Y.; Zhao, W.; Zhao, D.; Wang, C.; Yu, N.; An, T.; Mo, F.; Liu, J.; Miao, J.; Lv, B.; et al. Salvianolic Acid B Improves Mitochondrial Function in 3T3-L1 Adipocytes Through a Pathway Involving PPARγ Coactivator-1α (PGC-1α). Front. Pharmacol. 2018, 9, 671. [Google Scholar] [CrossRef]
- Lee, Y.G.; Park, D.H.; Chae, Y.C. Role of Mitochondrial Stress Response in Cancer Progression. Cells 2022, 11, 771. [Google Scholar] [CrossRef]
- Vafai, S.B.; Mootha, V.K. Mitochondrial disorders as windows into an ancient organelle. Nature 2012, 491, 374–383. [Google Scholar] [CrossRef] [PubMed]
- de Mello, A.H.; Costa, A.B.; Engel, J.D.G.; Rezin, G.T. Mitochondrial dysfunction in obesity. Life Sci. 2018, 192, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Hartman, M.-L.; Shirihai, O.S.; Holbrook, M.; Xu, G.; Kocherla, M.; Shah, A.; Fetterman, J.L.; Kluge, M.A.; Frame, A.A.; Hamburg, N.M. Relation of mitochondrial oxygen consumption in peripheral blood mononuclear cells to vascular function in type 2 diabetes mellitus. Vasc. Med. 2014, 19, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Bhatti, J.S.; Bhatti, G.K.; Reddy, P.H. Mitochondrial dysfunction and oxidative stress in metabolic disorders—A step towards mitochondria based therapeutic strategies. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2017, 1863, 1066–1077. [Google Scholar] [CrossRef]
- Tapondjou, L.A.; Ponou, K.B.; Teponno, R.B.; Mbiantcha, M.; Djoukeng, J.D.; Nguelefack, T.B.; Watcho, P.; Cadenas, A.G.; Park, H.J. In vivo anti-inflammatory effect of a new steroidal saponin, mannioside A, and its derivatives isolated from Dracaena mannii. Arch. Pharm. Res. 2008, 31, 653–658. [Google Scholar] [CrossRef]
- Zhu, L.; Tan, J.; Wang, B.; Guan, L.; Liu, Y.; Zheng, C. In-vitro Antitumor Activity and Antifungal Activity of Pennogenin Steroidal Saponins from paris Polyphylla var. yunnanensis. Iran. J. Pharm. Res. 2011, 10, 279–286. [Google Scholar]
- Fu, Y.L.; Yu, Z.Y.; Tang, X.M.; Zhao, Y.; Yuan, X.L.; Wang, S.; Ma, B.P.; Cong, Y.W. Pennogenin glycosides with a spirostanol structure are strong platelet agonists: Structural requirement for activity and mode of platelet agonist synergism. J. Thromb. Haemost. 2008, 6, 524–533. [Google Scholar] [CrossRef]
- Ur Rahman, S.; Adhikari, A.; Ismail, M.; Raza Shah, M.; Khurram, M.; Shahid, M.; Ali, F.; Haseeb, A.; Akbar, F.; Iriti, M. Beneficial Effects of Trillium govanianum Rhizomes in Pain and Inflammation. Molecules 2016, 21, 1095. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Park, S.K.; Lee, J.; Park, S.H.; Kim, Y.S.; Park, J.H.; Yu, S.; Lee, Y.G. Patulin Ameliorates Hypertrophied Lipid Accumulation and Lipopolysaccharide-Induced Inflammatory Response by Modulating Mitochondrial Respiration. Antioxidants 2023, 12, 1750. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Park, H.; Kim, W. Trehalose Inhibits Inflammatory Responses through Mitochondrial Reprogramming in RAW 264.7 Macrophages. Antioxidants 2023, 12, 1166. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, S.; Lee, H.M.; Lee, J.; Hwang, J.-T.; Choi, H.-K.; Lee, Y.G. Pennogenin 3-O-β-Chacotrioside Attenuates Hypertrophied Lipid Accumulation by Enhancing Mitochondrial Oxidative Capacity. Int. J. Mol. Sci. 2024, 25, 2970. https://doi.org/10.3390/ijms25052970
Yu S, Lee HM, Lee J, Hwang J-T, Choi H-K, Lee YG. Pennogenin 3-O-β-Chacotrioside Attenuates Hypertrophied Lipid Accumulation by Enhancing Mitochondrial Oxidative Capacity. International Journal of Molecular Sciences. 2024; 25(5):2970. https://doi.org/10.3390/ijms25052970
Chicago/Turabian StyleYu, Seungmin, Hee Min Lee, Jangho Lee, Jin-Taek Hwang, Hyo-Kyoung Choi, and Yu Geon Lee. 2024. "Pennogenin 3-O-β-Chacotrioside Attenuates Hypertrophied Lipid Accumulation by Enhancing Mitochondrial Oxidative Capacity" International Journal of Molecular Sciences 25, no. 5: 2970. https://doi.org/10.3390/ijms25052970
APA StyleYu, S., Lee, H. M., Lee, J., Hwang, J. -T., Choi, H. -K., & Lee, Y. G. (2024). Pennogenin 3-O-β-Chacotrioside Attenuates Hypertrophied Lipid Accumulation by Enhancing Mitochondrial Oxidative Capacity. International Journal of Molecular Sciences, 25(5), 2970. https://doi.org/10.3390/ijms25052970