Seasonal and Agricultural Response of Acidobacteria Present in Two Fynbos Rhizosphere Soils
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Data
2.2. Sequence Processing
2.3. Statistical Analysis
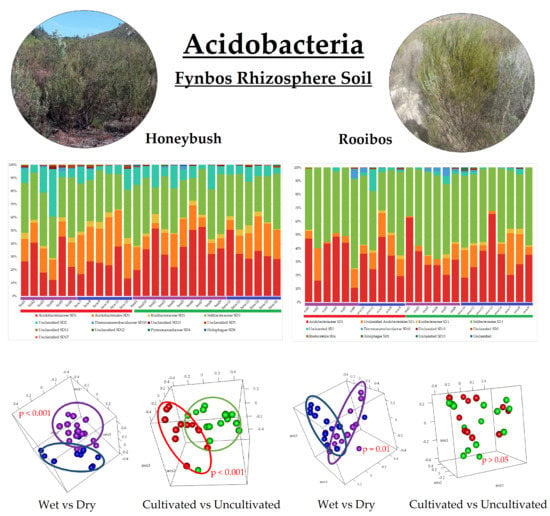
3. Results
3.1. Abiotic Soil Properties
3.2. Acidobacterial Community Composition
3.3. Alpha-Diversity
3.4. Beta-Diversity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rebelo, A.G.; Boucher, C.; Helme, N.; Mucina, L.; Rutherford, M.C. Fynbos Biome. In The Vegetation of South Africa, Lesotho and Swaziland; South African National Biodiversity Institute: Pretoria, South Africa, 2006; pp. 52–219. [Google Scholar]
- Cowling, R.M.; Rundel, P.W.; Lamont, B.B.; Arroyo, M.K.; Arianoutsou, M. Plant diversity in mediterranean-climate regions. Trends Ecol. Evol. 1996, 11, 362–366. [Google Scholar] [CrossRef]
- Cowling, R.M.; Ojeda, F.; Lamont, B.B.; Rundel, P.W.; Lechmere-Oertel, R. Rainfall reliability, a neglected factor in explaining convergence and divergence of plant traits in fire-prone mediterranean-climate ecosystems. Glob. Ecol. Biogeogr. 2005, 14, 509–519. [Google Scholar] [CrossRef]
- Joubert, E.; Gelderblom, W.C.A.; Louw, A.; de Beer, D. South African herbal teas: Aspalathus linearis, Cyclopia spp. and Athrixia phylicoides-A review. J. Ethnopharmacol. 2008, 119, 376–412. [Google Scholar] [CrossRef] [PubMed]
- Navarrete, A.A.; Kuramae, E.E.; de Hollander, M.; Pijl, A.S.; van Veen, J.A.; Tsai, S.M. Acidobacterial community responses to agricultural management of soybean in Amazon forest soils. FEMS Microbiol. Ecol. 2013, 83, 607–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montecchia, M.S.; Tosi, M.; Soria, M.A.; Vogrig, J.A.; Sydorenko, O.; Correa, O.S.; Moora, M. Pyrosequencing Reveals Changes in Soil Bacterial Communities after Conversion of Yungas Forests to Agriculture. PLoS ONE 2015, 10, e0119426. [Google Scholar] [CrossRef]
- Trivedi, P.; Delgado-Baquerizo, M.; Anderson, I.C.; Singh, B.K. Response of soil properties and microbial communities to agriculture: Implications for primary productivity and soil health indicators. Front. Plant Sci. 2016, 7, 990. [Google Scholar] [CrossRef] [Green Version]
- Jesus, E.D.C.; Marsh, T.L.; Tiedje, J.M.; Moreira, F.M.D.S. Changes in land use alter the structure of bacterial communities in Western Amazon soils. ISME J. 2009, 3, 1004–1011. [Google Scholar] [CrossRef]
- Fierer, N.; Lauber, C.L.; Ramirez, K.S.; Zaneveld, J.; Bradford, M.A.; Knight, R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012, 6, 1007–1017. [Google Scholar] [CrossRef] [Green Version]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef] [Green Version]
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef]
- Fierer, N.; Schimel, J.P. Effects of drying-rewetting frequency on soil carbon and nitrogen transformations. Soil Biol. Biochem. 2002, 34, 777–787. [Google Scholar] [CrossRef]
- Postma, A.; Slabbert, E.; Postma, F.; Jacobs, K. Soil bacterial communities associated with natural and commercial Cyclopia spp. FEMS Microbiol. Ecol. 2016, 92, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brink, C.; Postma, A.; Slabbert, E.; Postma, F.; Muasya, A.; Jacobs, K. Bacterial communities associated with natural and commercially grown rooibos (Aspalathus linearis). Pedosphere 2019, in press. [Google Scholar]
- Aulakh, M.S.; Wassmann, R.; Bueno, C.; Kreuzwieser, J.; Rennenberg, H. Characterization of root exudates at different growth stages of ten rice (Oryza sativa L.) cultivars. Plant Biol. 2001, 3, 139–148. [Google Scholar] [CrossRef] [Green Version]
- Bell, C.W.; Acosta-Martinez, V.; McIntyre, N.E.; Cox, S.; Tissue, D.T.; Zak, J.C. Linking microbial community structure and function to seasonal differences in soil moisture and temperature in a Chihuahuan Desert grassland. Microb. Ecol. 2009, 58, 827–842. [Google Scholar] [CrossRef]
- Pietikäinen, J.; Pettersson, M.; Bååth, E. Comparison of temperature effects on soil respiration and bacterial and fungal growth rates. FEMS Microbiol. Ecol. 2005, 52, 49–58. [Google Scholar] [CrossRef]
- Barns, S.M.; Takala, S.L.; Kuske, C.R. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl. Environ. Microbiol. 1999, 65, 1731–1737. [Google Scholar] [CrossRef] [Green Version]
- Tank, M.; Costas, A.M.G.; Bryant, D.A. Chloracidobacterium. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons Ltd.: New York, NY, USA, 2018; pp. 1–9. ISBN 9781118960608. [Google Scholar]
- Huber, K.J.; Geppert, A.M.; Wanner, G.; Fösel, B.U.; Wüst, P.K.; Overmann, J. The first representative of the globally widespread subdivision 6 Acidobacteria, Vicinamibacter silvestris gen. nov., sp. nov., isolated from subtropical savannah soil. Int. J. Syst. Evol. Microbiol. 2016, 66, 2971–2979. [Google Scholar] [CrossRef]
- Vieira, S.; Luckner, M.; Wanner, G.; Overmann, J. Luteitalea pratensis gen. nov., sp. nov. a new member of subdivision 6 Acidobacteria isolated from temperate grassland soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 1408–1414. [Google Scholar] [CrossRef]
- Quaiser, A.; López-García, P.; Zivanovic, Y.; Henn, M.R.; Rodriguez-Valera, F.; Moreira, D. Comparative analysis of genome fragments of Acidobacteria from deep Mediterranean plankton. Environ. Microbiol. 2008, 10, 2704–2717. [Google Scholar] [CrossRef]
- O’Connor-Sánchez, A.; Rivera-Domínguez, A.J.; de los Santos-Briones, C.; López-Aguiar, L.K.; Peña-Ramírez, Y.J.; Prieto-Davo, A. Acidobacteria appear to dominate the microbiome of two sympatric Caribbean Sponges and one Zoanthid. Biol. Res. 2014, 47, 67–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, J.; Gonzalez, J.M.; Saiz-Jimenez, C.; Ludwig, W. Detection and phylogenetic relationships of highly diverse uncultured acidobacterial communities in Altamira Cave using 23S rRNA sequence analyses. Geomicrobiol. J. 2005, 22, 379–388. [Google Scholar] [CrossRef]
- Hugenholtz, P.; Pitulle, C.; Hershberger, K.L.; Pace, N.R. Novel Division Level Bacterial Diversity in a Yellowstone Hot Spring Novel Division Level Bacterial Diversity in a Yellowstone Hot Spring. J. Bacteriol. 1998, 180, 366–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleinsteuber, S.; Müller, F.D.; Chatzinotas, A.; Wendt-Potthoff, K.; Harms, H. Diversity and in situ quantification of Acidobacteria subdivision 1 in an acidic mining lake. FEMS Microbiol. Ecol. 2008, 63, 107–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barns, S.M.; Cain, E.C.; Sommerville, L.; Kuske, C.R. Acidobacteria phylum sequences in uranium-contaminated subsurface sediments greatly expand the known diversity within the phylum. Appl. Environ. Microbiol. 2007, 73, 3113–3116. [Google Scholar] [CrossRef] [Green Version]
- Ludwig, W.; Bauer, S.H.; Bauer, M.; Held, I.; Kirchhof, G.; Huber, I.; Spring, S.; Hartmann, A.; Schleifer, K.H. Detection and in situ identification of representatives o fa widely distributed new bacterial phylum. FEMS Microbiol. Lett. 1997, 153, 181–190. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Yilmaz, P. Refining the taxonomic structure of the phylum Acidobacteria. Int. J. Syst. Evol. Microbiol. 2018, 68, 3796–3806. [Google Scholar] [CrossRef]
- Janssen, P.H. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 2006, 72, 1719–1728. [Google Scholar] [CrossRef] [Green Version]
- Jones, R.T.; Robeson, M.S.; Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. Int. Soc. Microb. Ecol. 2009, 3, 442–453. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Sinninghe Damsté, J.S. Acidobacteria. In eLS; John Wiley & Sons Ltd.: Chichester, UK, 2018; pp. 1–10. [Google Scholar]
- Rodrigues, J.L.M.; Pellizari, V.H.; Mueller, R.; Baek, K.; Jesus, E.D.C.; Paula, F.S.; Mirza, B.; Hamaou, G.S.; Tsai, S.M.; Feiglf, B.; et al. Conversion of the Amazon rainforest to agriculture results in biotic homogenization of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2013, 110, 988–993. [Google Scholar] [CrossRef] [Green Version]
- Sait, M.; Davis, K.E.R.; Janssen, P.H. Effect of pH on isolation and distribution of members of subdivision 1 of the phylum Acidobacteria occurring in soil. Appl. Environ. Microbiol. 2006, 72, 1852–1857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navarrete, A.A.; Venturini, A.M.; Meyer, K.M.; Klein, A.M.; Tiedje, J.M.; Brendan, B.J.; Nüsslein, K.; Tsai, S.M.; Rodrigues, J.L.M. Differential response of Acidobacteria subgroups to forest-to-pasture conversion and their biogeographic patterns in the western Brazilian Amazon. Front. Microbiol. 2015, 6, 1443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef] [PubMed]
- Kielak, A.; Pijl, A.S.; Van Veen, J.A.; Kowalchuk, G.A. Phylogenetic diversity of Acidobacteria in a former agricultural soil. ISME J. 2009, 3, 378–382. [Google Scholar] [CrossRef]
- Da Rocha, U.N.; Plugge, C.M.; George, I.; Van Elsas, J.D.; Van Overbeek, L.S. The rhizosphere selects for particular groups of Acidobacteria and Verrucomicrobia. PLoS ONE 2013, 8, e82443. [Google Scholar]
- Maseko, S.T.; Dakora, F.D. Accumulation of mineral elements in the rhizosphere and shoots of Cyclopia and Aspalathus species under different settings of the Cape fynbos. S. Afr. J. Bot. 2017, 110, 103–109. [Google Scholar] [CrossRef]
- Schloss, P.D.; Gevers, D.; Westcott, S.L. Reducing the Effects of PCR Amplification and Sequencing Artifacts on 16S rRNA-Based Studies. PLoS ONE 2011, 6, e27310. [Google Scholar] [CrossRef] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [Green Version]
- Keet, J.H.; Ellis, A.G.; Hui, C.; Le Roux, J.J. Strong spatial and temporal turnover of soil bacterial communities in South Africa’s hyperdiverse fynbos biome. Soil Biol. Biochem. 2019, 136, 107541. [Google Scholar] [CrossRef]
- Stafford, W.H.L.; Baker, G.C.; Brown, S.A.; Burton, S.G.; Cowan, D.A. Bacterial diversity in the rhizosphere of Proteaceae species. Environ. Microbiol. 2005, 7, 1755–1768. [Google Scholar] [CrossRef] [PubMed]
- Slabbert, E.; Jacobs, S.M.; Jacobs, K. The soil bacterial communities of South African fynbos riparian ecosystems invaded by Australian Acacia species. PLoS ONE 2014, 9, e86560. [Google Scholar] [CrossRef] [PubMed]
- Ward, N.L.; Challacombe, J.F.; Janssen, P.H.; Henrissat, B.; Coutinho, P.M.; Wu, M.; Xie, G.; Haft, D.H.; Sait, M.; Badger, J.; et al. Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microbiol. 2009, 75, 2046–2056. [Google Scholar] [CrossRef] [Green Version]
- Männistö, M.K.; Kurhela, E.; Tiirola, M.; Häggblom, M.M. Acidobacteria dominate the active bacterial communities of Arctic tundra with widely divergent winter-time snow accumulation and soil temperatures. FEMS Microbiol. Ecol. 2013, 84, 47–59. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Cho, J.C. Distribution patterns of the members of phylum Acidobacteria in global soil samples. J. Microbiol. Biotechnol. 2009, 19, 1281–1287. [Google Scholar] [CrossRef]
- Mikha, M.M.; Rice, C.W.; Milliken, G.A. Carbon and nitrogen mineralization as affected by drying and wetting cycles. Soil Biol. Biochem. 2005, 37, 339–347. [Google Scholar] [CrossRef]
- Van Gestel, M.; Ladd, J.N.; Amato, M. Microbial biomass responses to seasonal change and imposed drying regimes at increasing depths of undisturbed topsoil profiles. Soil Biol. Biochem. 1992, 24, 103–111. [Google Scholar] [CrossRef]
- Castro, H.F.; Classen, A.T.; Austin, E.E.; Norby, R.J.; Schadt, C.W. Soil Microbial Community Responses to Multiple Experimental Climate Change Drivers. Appl. Environ. Microbiol. 2010, 76, 999–1007. [Google Scholar] [CrossRef] [Green Version]
- Lauber, C.L.; Ramirez, K.S.; Aanderud, Z.; Lennon, J.; Fierer, N. Temporal variability in soil microbial communities across land-use types. ISME J. 2013, 7, 1641–1650. [Google Scholar] [CrossRef]
- de Menezes, A.B.; Müller, C.; Clipson, N.; Doyle, E. The soil microbiome at the Gi-FACE experiment responds to a moisture gradient but not to CO2 enrichment. Microbiology 2016, 162, 1572–1582. [Google Scholar] [CrossRef]
- Lee, K.C.; Caruso, T.; Archer, S.D.J.; Gillman, L.N.; Lau, M.C.Y.; Cary, S.C.; Lee, C.K.; Pointing, S.B. Stochastic and Deterministic Effects of a Moisture Gradient on Soil Microbial Communities in the McMurdo Dry Valleys of Antarctica. Front. Microbiol. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bardgett, R.D.; Freeman, C.; Ostle, N.J. Microbial contributions to climate change through carbon cycle feedbacks. ISME J. 2008, 2, 805–814. [Google Scholar] [CrossRef] [Green Version]
- Parton, W.; Silver, W.L.; Burke, I.C.; Grassens, L.; Harmon, M.E.; Currie, W.S.; King, J.Y.; Carol Adair, E.; Brandt, L.A.; Hart, S.C.; et al. Global-Scale Similarities in Nitrogen Release Patterns During Long-Term Decomposition. Science 2007, 315, 361–364. [Google Scholar] [CrossRef] [PubMed]
- Pankratov, T.A.; Dedysh, S.N. Granulicella paludicola gen. nov., sp. nov., Granulicella pectinivorans sp. nov., Granulicella aggregans sp. nov. and Granulicella rosea sp. nov., acidophilic, polymer-degrading acidobacteria from Sphagnum peat bogs. Int. J. Syst. Evol. Microbiol. 2010, 60, 2951–2959. [Google Scholar] [CrossRef] [PubMed]
- Männistö, M.K.; Rawat, S.; Starovoytov, V.; Häggblom, M.M. Terriglobus saanensis sp. nov., an acidobacterium isolated from tundra soil. Int. J. Syst. Evol. Microbiol. 2011, 61, 1823–1828. [Google Scholar] [CrossRef] [PubMed]
- García-Fraile, P.; Benada, O.; Cajthaml, T.; Baldrian, P.; Llado, S. Terracidiphilus gabretensis gen. nov., sp. nov., an Abundant and Active Forest Soil Acidobacterium Important in Organic Matter Transformation. Appl. Environ. Microbiol. 2016, 82, 560–569. [Google Scholar] [CrossRef] [Green Version]
- Denef, K.; Six, J.; Bossuyt, H.; Frey, S.D.; Elliott, E.T.; Merckx, R.; Paustian, K. Influence of dry-wet cycles on the interrelationship between aggregate, particulate organic matter, and microbial community dynamics. Soil Biol. Biochem. 2001, 33, 1599–1611. [Google Scholar] [CrossRef]
- Halverson, L.J.; Jones, T.M.; Firestone, M.K. Release of Intracellular Solutes by Four Soil Bacteria Exposed to Dilution Stress. Soil Sci. Soc. Am. J. 2000, 64, 1630. [Google Scholar] [CrossRef]
- Pereira de Castro, A.; Sartori da Silva, M.R.S.; Quirino, B.F.; da Cunha Bustamante, M.M.; Krüger, R.H. Microbial Diversity in Cerrado Biome (Neotropical Savanna) Soils. PLoS ONE 2016, 11, e0148785. [Google Scholar] [CrossRef] [Green Version]
- Catão, E.C.P.P.; Lopes, F.A.C.C.; Araújo, J.F.; De Castro, A.P.; Barreto, C.C.; Bustamante, M.M.C.C.; Quirino, B.F.; Krüger, R.H.; Barreto, C.C.; Krüger, R.H.; et al. Soil Acidobacterial 16S rRNA Gene Sequences Reveal Subgroup Level Differences between Savanna-Like Cerrado and Atlantic Forest Brazilian Biomes. Int. J. Microbiol. 2014, 2014, 1–12. [Google Scholar] [CrossRef]
- Challacombe, J.F.; Eichorst, S.A.; Hauser, L.; Land, M.; Xie, G. Biological Consequences of Ancient Gene Acquisition and Duplication in the Large Genome of Candidatus Solibacter usitatus Ellin6076. PLoS ONE 2011, 6, 24882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kielak, A.M.; Barreto, C.C.; Kowalchuk, G.A.; van Veen, J.A.; Kuramae, E.E. The ecology of Acidobacteria: Moving beyond genes and genomes. Front. Microbiol. 2016, 7, 744. [Google Scholar] [CrossRef] [Green Version]
- Singh, B.K.; Munro, S.; Potts, J.M.; Millard, P. Influence of grass species and soil type on rhizosphere microbial community structure in grassland soils. Appl. Soil Ecol. 2007, 36, 147–155. [Google Scholar] [CrossRef]
- Lee, S.H.; Ka, J.O.; Cho, J.C. Members of the phylum Acidobacteria are dominant and metabolically active in rhizosphere soil. FEMS Microbiol. Lett. 2008, 285, 263–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalam, S.; Das, S.N.; Basu, A.; Podile, A.R. Population densities of indigenous Acidobacteria change in the presence of plant growth promoting rhizobacteria (PGPR) in rhizosphere. J. Basic Microbiol. 2017, 57, 376–385. [Google Scholar] [CrossRef]
- Crits-Christoph, A.; Diamond, S.; Butterfield, C.N.; Thomas, B.C.; Banfield, J.F. Novel soil bacteria possess diverse genes for secondary metabolite biosynthesis. Nature 2018, 558, 440–444. [Google Scholar] [CrossRef]
- Liu, J.; Sui, Y.; Yu, Z.; Yao, Q.; Shi, Y.; Chu, H.; Jin, J.; Liu, X.; Wang, G. Diversity and distribution patterns of acidobacterial communities in the black soil zone of northeast China. Soil Biol. Biochem. 2016, 95, 212–222. [Google Scholar] [CrossRef]
- Li, W.; Liu, X.; Niu, S. Differential responses of the acidobacterial community in the topsoil and subsoil to fire disturbance in Pinus tabulaeformis stands. PeerJ 2019, 7, e8047. [Google Scholar] [CrossRef] [Green Version]
- Sui, X.; Zhang, R.; Frey, B.; Yang, L.; Li, M.H.; Ni, H. Land use change effects on diversity of soil bacterial, Acidobacterial and fungal communities in wetlands of the Sanjiang Plain, northeastern China. Sci. Rep. 2019, 9, 18535. [Google Scholar] [CrossRef]





| Soil Properties | Dry 1 | Wet 1 | Cultivated 2 | Uncultivated 2 |
|---|---|---|---|---|
| pH | 4.15 ± 0.67 | 4.37 ± 0.61 | 4.75 ± 0.42 * | 3.83 ± 0.46 * |
| H+ | 3.03 ± 2.61 | 2.31 ± 2.11 | 1.42 ± 1.40 | 3.80 ± 2.55 |
| P | 3.76 ± 2.63 | 5.00 ± 2.63 | 5.23 ± 2.83 | 3.50 ± 2.31 |
| K+ | 52.18 ± 14.23 | 69.17 ± 33.29 | 56.69 ± 28.75 | 61.25 ± 22.20 |
| Na+ | 0.18 ± 0.07 | 0.17 ± 0.09 | 0.12 ± 0.04 | 0.21 ± 0.08 |
| Ca2+ | 1.88 ± 0.87 | 1.51 ± 0.79 | 1.41 ± 0.77 | 1.99 ± 0.84 |
| Mg2+ | 1.27 ± 0.65 | 1.03 ± 0.63 | 0.92 ± 0.58 * | 1.38 ± 0.63 * |
| C | 2.05 ± 0.73 * | 11.31 ± 4.90 * | 6.88 ± 5.74 | 5.08 ± 5.52 |
| NO3-N | 1.28 ± 1.27 | 1.48 ± 1.78 | 2.26 ± 1.85 * | 0.63 ± 0.20 * |
| NH4-N | 7.08 ± 1.50 | 7.59 ± 1.38 | 7.28 ± 1.75 | 7.30 ± 1.21 |
| Soil Properties | Dry 1 | Wet 1 | Cultivated 2 | Uncultivated 2 |
|---|---|---|---|---|
| pH | 4.65 ± 0.38 | 4.48 ± 0.29 | 4.29 ± 0.24 * | 4.81 ± 0.19 * |
| H+ | 0.57 ± 0.30 | 0.50 ± 0.22 | 0.58 ± 0.30 | 0.49 ± 0.22 |
| P | 6.67 ± 2.10 | 5.54 ± 5.06 | 8.00 ± 4.13 * | 4.31 ± 2.75 * |
| K+ | 49.58 ± 34.08 | 24.62 ± 9.00 | 48.42 ± 34.26 | 25.69 ± 11.18 |
| Na+ | 0.10 ± 0.07 | 0.05 ± 0.03 | 0.09 ± 0.08 | 0.06 ± 0.03 |
| Ca2+ | 0.65 ± 0.36 | 0.81 ± 0.54 | 0.40 ± 0.20 | 1.04 ± 0.41 |
| Mg2+ | 0.32 ± 0.05 | 0.34 ± 0.24 | 0.25 ± 0.08 | 0.41 ± 0.20 |
| C | 0.31 ± 0.15 * | 0.77 ± 0.60 * | 0.39 ± 0.20 | 0.70 ± 0.64 |
| NO3-N | 1.30 ± 0.75 | 0.84 ± 0.72 | 1.49 ± 0.87 | 0.66 ± 0.29 |
| NH4-N | 7.76 ± 1.83 | 8.01 ± 0.85 | 8.09 ± 1.57 | 7.71 ± 1.21 |
| Group | Group ID | Filtered Reads 1 | Acidobacteria Reads | Average Relative Abundance (%) | p-Value 2 |
|---|---|---|---|---|---|
| Honeybush Cultivated | HC | 238,526 | 22,518 | 9.4 ± 5.1 | 0.09 |
| Honeybush Uncultivated | HN | 405,815 | 28,270 | 7.0 ± 2.4 | |
| Honeybush Dry | HD | 451,358 | 31,591 | 7.0 ± 2.5 | 0.19 |
| Honeybush Wet | HW | 192,983 | 19,197 | 9.9 ± 4.9 | |
| Rooibos Cultivated | RC | 514,844 | 21,691 | 4.2 ± 5.4 | 0.14 |
| Rooibos Uncultivated | RN | 521,043 | 23,498 | 4.5 ± 3.9 | |
| Rooibos Dry | RD | 717,981 | 14,811 | 2.1 ± 2.0 | 0.006 * |
| Rooibos Wet | RW | 317,906 | 30,378 | 9.6 ± 2.8 |
| Taxonomic Classification | HC | HN | HW | HD | RC | RN | RD | RW | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subdivision | Family | Genus | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD |
| SD1 | Acidobacteriaceae | Acidicapsa | 1.3 | 1.5 | 2.2 | 1.8 | 1.2 | 1.5 | 2.2 | 1.7 | 0.5 | 0.6 | 0.3 | 1.0 | 0.5 | 1.1 | 0.3 | 0.6 |
| Acidipila | 3.0 * | 2.0 | 9.4 * | 5.0 | 4.6 * | 5.0 | 7.5 * | 5.3 | 8.3 | 9.0 | 7.7 | 5.2 | 9.1 | 8.7 | 7.5 | 5.1 | ||
| Unclassified | 0.1 | 0.2 | 0.1 | 0.2 | 0.1 | 0.2 | 0.1 | 0.2 | 0.1 | 0.1 | 0.2 | 0.4 | 0.2 | 0.2 | 0.1 | 0.4 | ||
| Unclassified | 10.1 | 5.5 | 13.8 | 6.0 | 11.5 | 5.2 | 12.5 | 6.3 | 12.8 | 6.3 | 11.6 | 8.9 | 9.6 * | 5.4 | 13.1 * | 9.2 | ||
| Bryocella | 0.1 * | 0.1 | 0.4 * | 1.0 | 0.4 | 1.1 | 0.2 | 0.3 | 0.6 | 3.5 | 0.8 | 1.2 | 1.0 | 3.4 | 0.6 | 1.2 | ||
| Edaphobacter | 2.3 | 2.4 | 1.4 | 1.8 | 1.5 | 1.2 | 2.0 | 2.6 | 1.5 | 2.1 | 1.4 | 2.1 | 2.3 | 2.8 | 1.1 | 0.8 | ||
| Granulicella | 2.1 * | 0.9 | 4.4 * | 3.3 | 2.7 * | 1.9 | 3.7 * | 3.5 | 4.3 | 2.6 | 4.1 | 4.3 | 5.1 | 4.3 | 3.8 | 2.8 | ||
| Occallatibacter | 1.3 | 1.0 | 1.4 | 0.9 | 1.5 | 0.8 | 1.2 | 1.0 | 0.6 | 0.7 | 1.0 | 1.8 | 0.9 | 2.0 | 0.8 | 0.6 | ||
| Telmatobacter | - | - | - | - | - | - | <0.1 | 0.1 | - | - | - | - | <0.1 | 0.1 | ||||
| Terracidiphilus | 2.2 | 1.3 | 1.6 | 1.0 | 1.7 | 1.2 | 1.9 | 1.3 | 1.4 | 1.2 | 1.3 | 1.4 | 1.0 | 1.7 | 1.5 | 0.8 | ||
| Terriglobus | 0.6 | 1.7 | 0.3 | 0.5 | 0.4 | 0.4 | 0.5 | 1.5 | 0.2 | 0.3 | 0.2 | 0.4 | <0.1 | 0.4 | 0.3 | 0.3 | ||
| Unclassified | 0.9 | 0.6 | 0.4 | 0.5 | 1.1 * | 0.6 | 0.3 * | 0.4 | 1.0 | 0.6 | 1.4 | 1.1 | 0.6 | 0.5 | 1.4 | 0.9 | ||
| Unclassified | Unclassified | 0.1 | 0.2 | - | - | - | <0.1 | 0.2 | - | - | 0.1 | 0.1 | - | - | <0.1 | 0.1 | ||
| Unclassified | Unclassified | 2.5 | 1.1 | 1.6 | 2.0 | 2.6 | 1.9 | 1.7 | 1.5 | 3.0 | 1.7 | 5.3 | 4.0 | 2.3 | 1.7 | 5.0 | 3.8 | |
| Koribacteraceae | Candidatus Koribacter | 2.9 | 1.4 | 2.7 | 1.9 | 3.3 | 1.8 | 2.5 | 1.6 | 1.6 | 1.0 | 2.0 | 1.3 | 1.1 | 0.9 | 2.1 | 1.0 | |
| Unclassified | Unclassified | 20.3 | 7.2 | 13.7 | 6.7 | 23.2 * | 6.9 | 13.1 * | 5.4 | 8.9 | 5.6 | 8.9 | 5.1 | 6.8 | 5.4 | 9.7 | 5.0 | |
| Unclassified | Unclassified | 0.1 | 0.1 | - | - | 0.1 | 0.1 | - | - | 0.1 | 0.1 | 0.1 | 0.1 | <0.1 | 0.1 | 0.1 | 0.1 | |
| SD2 | Unclassified | Unclassified | 13.2 | 9.2 | 9.7 | 4.6 | 9.0 * | 3.5 | 12.7 * | 8.3 | 3.9 | 4.7 | 1.9 | 1.7 | 1.7 | 1.8 | 3.2 | 4.2 |
| SD3 | Solibacteraceae | Unclassified | 0.1 | 0.3 | - | - | 0.1 | 0.3 | - | - | - | - | - | - | - | - | - | |
| Bryobacter | 13.3 | 1.8 | 14.8 | 4.2 | 14.2 | 3.9 | 14.0 | 3.1 | 23.2 | 10.4 | 21.0 | 7.3 | 23.8 | 10.6 | 21.3 | 6.8 | ||
| Candidatus Solibacter | 18.0 | 4.7 | 16.4 | 6.7 | 16.5 | 4.0 | 17.5 | 7.2 | 16.6 | 8.4 | 16.8 | 10.8 | 19.8 | 13.4 | 15.5 | 5.4 | ||
| Unclassified | <0.1 | 0.1 | - | - | <0.1 | 0.1 | - | - | - | - | <0.1 | 0.1 | - | - | <0.1 | 0.1 | ||
| Paludibaculum | <0.1 | 0.1 | <0.1 | 0.1 | <0.1 | 0.1 | <0.1 | 0.1 | - | - | 0.1 | 0.2 | 0.1 | 0.2 | - | - | ||
| Unclassified | 3.4 | 1.5 | 4.7 | 3.0 | 3.0 | 2.1 | 4.8 | 2.7 | 9.5 | 5.9 | 12.0 | 4.9 | 10.4 | 5.7 | 11.1 | 4.9 | ||
| Unclassified | <0.1 | 0.1 | - | - | - | - | <0.1 | 0.1 | <0.1 | 0.0 | <0.1 | 0.1 | - | - | <0.1 | 0.1 | ||
| SD4 | Pyrinomonadaceae | Pyrinomonas | - | - | <0.1 | 0.2 | <0.1 | 0.2 | - | - | - | - | - | - | - | - | - | - |
| Unclassified | Unclassified | - | - | - | - | - | - | - | - | 0.1 | 0.3 | <0.1 | 0.1 | 0.1 | 0.3 | <0.1 | 0.1 | |
| SD5 | Unclassified | Unclassified | 0.9 * | 0.8 | 0.2 * | 0.4 | 0.7 | 0.7 | 0.4 | 0.7 | 0.4 | 0.4 | 0.3 | 0.3 | 0.4 | 0.4 | 0.3 | 0.3 |
| SD8 | Unclassified | Unclassified | 0.1 | 0.2 | 0.1 | 0.3 | 0.1 | 0.2 | 0.1 | 0.3 | <0.1 | 0.1 | 0.3 | 0.5 | 0.1 | 0.1 | 0.2 | 0.5 |
| SD10 | Thermoanaerobaculaceae | Unclassified | 0.6 | 0.8 | 0.3 | 0.7 | 0.2 * | 0.3 | 0.5 * | 0.9 | 1.3 | 2.3 | 1.2 | 1.6 | 2.6 | 2.5 | 0.7 | 1.0 |
| SD11 | Unclassified | Unclassified | <0.1 | 0.1 | - | - | - | - | <0.1 | 0.1 | - | - | - | - | - | - | - | - |
| SD12 | Unclassified | Unclassified | 0.1 | 0.1 | 0.1 | 0.3 | 0.1 | 0.1 | 0.1 | 0.3 | - | - | - | - | - | - | - | - |
| SD13 | Unclassified | Unclassified | 0.5 | 0.6 | 0.3 | 0.4 | 0.4 | 0.4 | 0.4 | 0.5 | <0.1 | 0.1 | 0.1 | 0.1 | - | - | 0.1 | 0.2 |
| SD15 | Unclassified | Unclassified | 0.1 | 0.2 | <0.1 | 0.1 | 0.1 | 0.2 | <0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | <0.1 | 0.0 | 0.1 | 0.2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Conradie, T.; Jacobs, K. Seasonal and Agricultural Response of Acidobacteria Present in Two Fynbos Rhizosphere Soils. Diversity 2020, 12, 277. https://doi.org/10.3390/d12070277
Conradie T, Jacobs K. Seasonal and Agricultural Response of Acidobacteria Present in Two Fynbos Rhizosphere Soils. Diversity. 2020; 12(7):277. https://doi.org/10.3390/d12070277
Chicago/Turabian StyleConradie, Tersia, and Karin Jacobs. 2020. "Seasonal and Agricultural Response of Acidobacteria Present in Two Fynbos Rhizosphere Soils" Diversity 12, no. 7: 277. https://doi.org/10.3390/d12070277
APA StyleConradie, T., & Jacobs, K. (2020). Seasonal and Agricultural Response of Acidobacteria Present in Two Fynbos Rhizosphere Soils. Diversity, 12(7), 277. https://doi.org/10.3390/d12070277