A Comparative Phylogeographic Approach to Facilitate Recovery of an Imperiled Freshwater Mussel (Bivalvia: Unionida: Potamilus inflatus)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Taxon Sampling
2.2. Phylogenetic Analyses
2.3. Phylogeographic Analyses
2.4. Comparative Phylogeography
3. Results
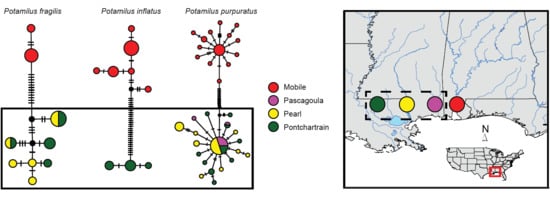
Molecular Analyses
4. Discussion
4.1. Patterns of Genetic Variation in Potamilus Species
4.2. Implications on Conservation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Butchart, S.H.M.; Walpole, M.; Collen, B.; van Strien, A.; Scharlemann, J.P.W.; Almond, R.E.A.; Baillie, J.E.M.; Bomhard, B.; Brown, C.; Bruno, J.; et al. Global biodiversity: Indicators of recent declines. Science 2010, 328, 1164–1168. [Google Scholar] [CrossRef]
- Rands, M.R.W.; Adams, W.M.; Bennun, L.; Butchart, S.H.M.; Clements, A.; Coomes, D.; Entwistle, A.; Hodge, I.; Kapos, V.; Scharlemann, J.P.W.; et al. Biodiversity conservation: Challenges beyond 2010. Science 2010, 329, 1298–1303. [Google Scholar] [CrossRef] [Green Version]
- Dudgeon, D.; Arthington, A.H.; Gessner, M.O.; Kawabata, Z.-I.; Knowler, D.J.; Lévêque, C.; Naiman, R.J.; Prieur-Richard, A.-H.; Soto, D.; Stiassny, M.L.J.; et al. Freshwater biodiversity: importance, threats, status and conservation challenges. Biol. Rev. 2006, 81, 163–182. [Google Scholar] [CrossRef]
- Williams, J.D.; Warren, M.L.; Cummings, K.S.; Harris, J.L.; Neves, R.J. Conservation status of freshwater mussels of the United States and Canada. Fisheries 1993, 18, 6–22. [Google Scholar] [CrossRef]
- Strayer, D.L.; Downing, J.A.; Haag, W.R.; King, T.L.; Layer, J.B.; Newton, T.J.; Nichols, S.J. Changing perspectives on pearly mussels, North America’s most imperiled animals. BioScience 2004, 54, 429–439. [Google Scholar] [CrossRef]
- Haag, W.R.; Williams, J.D. Biodiversity on the brink: An assessment of conservation strategies for North American freshwater mussels. Hydrobiologia 2014, 735, 45–60. [Google Scholar] [CrossRef]
- Watters, G.T. Form and function of unionoidean shell sculpture and shape (Bivalvia). Am. Malacol. Bull. 1993, 11, 1–20. [Google Scholar]
- Bringolf, R.B.; Cope, W.G.; Barnhart, M.C.; Mosher, S.; Lazaro, P.R.; Shea, D. Acute and chronic toxicity of pesticide formulations (atrazine, chlorpyrifos, and permethrin) to glochidia and juveniles of Lampsilis siliquoidea. Environ. Toxicol. Chem. 2007, 26, 2101–2107. [Google Scholar] [CrossRef]
- Wang, N.; Ingersoll, C.G.; Greer, I.E.; Hardesty, D.K.; Ivey, C.D.; Kunz, J.L.; Brumbaugh, W.G.; Dwyer, F.J.; Roberts, A.D.; Augspurger, T.; et al. Chronic toxicity of copper and ammonia to juvenile freshwater mussels (Unionidae). Environ. Toxicol. Chem. 2007, 26, 2048–2056. [Google Scholar] [CrossRef] [Green Version]
- Vaughn, C.C.; Taylor, C.M. Impoundments and the decline of freshwater mussels: A case study of an extinction gradient. Conserv. Biol. 1999, 13, 912–920. [Google Scholar] [CrossRef]
- Ferreira-Rodríguez, N.; Akiyama, Y.B.; Aksenova, O.V.; Araujo, R.; Christopher Barnhart, M.; Bespalaya, Y.V.; Bogan, A.E.; Bolotov, I.N.; Budha, P.B.; Clavijo, C.; et al. Research priorities for freshwater mussel conservation assessment. Biol. Conserv. 2019, 231, 77–87. [Google Scholar] [CrossRef] [Green Version]
- Allendorf, F.W.; Luikart, G.; Aitken, S.N. Conservation and the Genetics of Populations, 2nd ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2013; ISBN 978-0-470-67145-0. [Google Scholar]
- Unmack, P.J. Biogeography of Australian freshwater fishes. J. Biogeogr. 2001, 28, 1053–1089. [Google Scholar] [CrossRef]
- Oaks, J.R. An improved approximate-Bayesian model-choice method for estimating shared evolutionary history. BMC Evol. Biol. 2014, 14, 150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moritz, C.; Faith, D.P. Comparative phylogeography and the identification of genetically divergent areas for conservation. Mol. Ecol. 1998, 7, 419–429. [Google Scholar] [CrossRef]
- Hickerson, M.J.; Carstens, B.C.; Cavender-Bares, J.; Crandall, K.A.; Graham, C.H.; Johnson, J.B.; Rissler, L.; Victoriano, P.F.; Yoder, A.D. Phylogeography’s past, present, and future: 10 years after Avise, 2000. Mol. Phylogenet. Evol. 2010, 54, 291–301. [Google Scholar] [CrossRef]
- Bermingham, E.; Avise, J.C. Molecular zoogeography of freshwater fishes in the southeastern United States. Genetics 1986, 113, 939–965. [Google Scholar]
- Avise, J.C. Molecular population structure and the biogeographic history of a regional fauna: A case history with lessons for conservation biology. Oikos 1992, 63, 62–76. [Google Scholar] [CrossRef] [Green Version]
- Walker, D.; Avise, J.C. Principles of phylogeography as illustrated by freshwater and terrestrial turtles in the southeastern United States. Annu. Rev. Ecol. Syst. 1998, 29, 23–58. [Google Scholar] [CrossRef] [Green Version]
- Grantham, H.S.; Pressey, R.L.; Wells, J.A.; Beattie, A.J. Effectiveness of biodiversity surrogates for conservation planning: Different measures of effectiveness generate a kaleidoscope of variation. PLoS ONE 2010, 5, e11430. [Google Scholar] [CrossRef]
- Stewart, D.R.; Underwood, Z.E.; Rahel, F.J.; Walters, A.W. The effectiveness of surrogate taxa to conserve freshwater biodiversity. Conserv. Biol. 2018, 32, 183–194. [Google Scholar] [CrossRef]
- Galbraith, H.S.; Zanatta, D.T.; Wilson, C.C. Comparative analysis of riverscape genetic structure in rare, threatened and common freshwater mussels. Conserv. Genet. 2015, 16, 845–857. [Google Scholar] [CrossRef]
- Elderkin, C.L.; Christian, A.D.; Metcalfe-Smith, J.L.; Berg, D.J. Population genetics and phylogeography of freshwater mussels in North America, Elliptio dilatata and Actinonaias ligamentina (Bivalvia: Unionidae). Mol. Ecol. 2008, 17, 2149–2163. [Google Scholar] [CrossRef]
- Chong, J.P.; Roe, K.J. A comparison of genetic diversity and population structure of the endangered scaleshell mussel (Leptodea leptodon), the fragile papershell (Leptodea fragilis) and their host-fish the freshwater drum (Aplodinotus grunniens). Conserv. Genet. 2018, 19, 425–437. [Google Scholar] [CrossRef] [Green Version]
- Mock, K.E.; Brim Box, J.C.; Chong, J.P.; Furnish, J.; Howard, J.K. Comparison of population genetic patterns in two widespread freshwater mussels with contrasting life histories in western North America. Mol. Ecol. 2013, 22, 6060–6073. [Google Scholar] [CrossRef]
- Jones, J.W.; Neves, R.J.; Hallerman, E.M. Historical demography of freshwater mussels (Bivalvia: Unionidae): Genetic evidence for population expansion and contraction during the late Pleistocene and Holocene. Biol. J. Linn. Soc. 2015, 114, 376–397. [Google Scholar] [CrossRef] [Green Version]
- Williams, J.D.; Bogan, A.E.; Butler, R.S.; Cummings, K.S.; Garner, J.T.; Harris, J.L.; Johnson, N.A.; Watters, G.T. A revised list of the freshwater mussels (Mollusca: Bivalvia: Unionida) of the United States and Canada. Freshw. Mollusk Biol. Conserv. 2017, 20, 33–58. [Google Scholar] [CrossRef] [Green Version]
- Smith, C.H.; Pfeiffer, J.M.; Johnson, N.A. Comparative phylogenomics reveal complex evolution of life history strategies in a clade of bivalves with parasitic larvae (Bivalvia: Unionoida: Ambleminae). Cladistics 2020, in press. [Google Scholar] [CrossRef]
- Haag, W.R. North American Freshwater Mussels: Natural History, Ecology, and Conservation; Cambridge University Press: Cambridge, MA, USA, 2012; ISBN 978-0-521-19938-4. [Google Scholar]
- USFWS. Endangered and threatened wildlife and plants; determination of threatened status for the Inflated Heelsplitter, Potamilus inflatus. Fed. Regist. 1990, 55, 39868–39872. [Google Scholar]
- Williams, J.D.; Bogan, A.E.; Garner, J.T. Freshwater Mussels of Alabama and the Mobile Basin in Georgia; University of Alabama Press: Tuscaloosa, AL, USA, 2008; ISBN 978-0-8173-1613-6. [Google Scholar]
- Jones, R.L.; Wagner, M.D.; Slack, W.T.; Peyton, J.S.; Hartfield, P.D. Guide to the Identification and Distribution of Freshwater Mussels (Bivalvia: Unionidae) in Mississippi; Mississippi Department of Wildlife, Fisheries, and Parks: Jackson, MS, USA, 2019.
- Hartfield, P.D. Status Survey for the Alabama Heelsplitter Mussel Potamilus Inflatus (Lea, 1831); U.S. Fish and Wildlife Service: Jackson, MS, USA, 1988.
- Brown, K.M.; Daniel, W.M. The population ecology of the threatened Inflated Heelsplitter, Potamilus inflatus, in the Amite River, Louisiana. Am. Midl. Nat. 2014, 171, 328–339. [Google Scholar] [CrossRef]
- USFWS. Inflated Heelsplitter Mussel (Potamilus Inflatus) 5-Year Review: Summary and Evaluation; US Fish and Wildlife Service: Daphne, AL, USA, 2014.
- Frierson, L.S. A comparison of the Unionidae of the Pearl and Sabine rivers. Nautilus 1911, 24, 134–136. [Google Scholar]
- George, S.G.; Reine, K.J. Rediscovery of the Inflated Heelsplitter mussel, Potamilus inflatus, from the Pearl River drainage. J. Freshw. Ecol. 1996, 11, 245–246. [Google Scholar] [CrossRef]
- Brown, K.M.; Daniel, W.; George, G. The effect of Hurricane Katrina on the mussel assemblage of the Pearl River, Louisiana. Aquat. Ecol. 2010, 44, 223–231. [Google Scholar] [CrossRef]
- Brown, K.M.; Banks, P.D. The conservation of unionid mussels in Louisiana rivers: Diversity, assemblage composition and substrate use. Aquat. Conserv. 2001, 11, 189–198. [Google Scholar] [CrossRef]
- Brown, K.M.; Daniel, W.M. Mussel mortality from a toxic spill in the Pearl River, Louisiana. Ellipsaria 2012, 14, 28–31. [Google Scholar]
- LDWF. Investigation of a Fish and Mollusk Kill in the Lower Pearl River, Louisiana and Mississippi; Louisiana Department of Wildlife and Fisheries: Baton Rouge, LA, USA, 2011.
- Roe, K.J.; Lydeard, C. Molecular systematics of the freshwater mussel genus Potamilus (Bivalvia: Unionidae). Malacologia 1998, 39, 195–205. [Google Scholar]
- Pfeiffer, J.M.; Breinholt, J.W.; Page, L.M. Unioverse: Phylogenomic resources for reconstructing the evolution of freshwater mussels (Unionoida). Mol. Phylogenet. Evol. 2019, 137, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Johnson, N.A.; Smith, C.H. Novel genetic resources to facilitate future molecular studies in freshwater mussels (Bivalvia: Unionidae). Data. under review.
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Maddison, W.P.; Maddison, D.R. Mesquite: A modular system for evolutionary analysis. Version 3.31. 2017. Available online: https://www.mesquiteproject.org/ (accessed on 1 July 2020).
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chernomor, O.; von Haeseler, A.; Minh, B.Q. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016, 65, 997–1008. [Google Scholar] [CrossRef] [Green Version]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [Green Version]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Quang Minh, B.; Sy Vinh, L. Ufboot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Sukumaran, J.; Knowles, L.L. Multispecies coalescent delimits structure, not species. Proc. Natl. Acad. Sci. USA 2017, 114, 1607–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, C.H.; Johnson, N.A.; Pfeiffer, J.M.; Gangloff, M.M. Molecular and morphological data reveal non-monophyly and speciation in imperiled freshwater mussels (Anodontoides and Strophitus). Mol. Phylogenet. Evol. 2018, 119, 50–62. [Google Scholar] [CrossRef]
- Smith, C.H.; Johnson, N.A.; Inoue, K.; Doyle, R.D.; Randklev, C.R. Integrative taxonomy reveals a new species of freshwater mussel, Potamilus streckersoni sp. nov. (Bivalvia: Unionidae): Implications for conservation and management. Syst. Biodivers. 2019, 17, 331–348. [Google Scholar] [CrossRef] [Green Version]
- Pfeiffer, J.M.; Johnson, N.A.; Randklev, C.R.; Howells, R.G.; Williams, J.D. Generic reclassification and species boundaries in the rediscovered freshwater mussel ‘Quadrula’ mitchelli (Simpson in Dall, 1896). Conserv. Genet. 2016, 17, 279–292. [Google Scholar] [CrossRef]
- Jones, G. Algorithmic improvements to species delimitation and phylogeny estimation under the multispecies coalescent. J. Math. Biol. 2017, 74, 447–467. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.-H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef] [Green Version]
- Heled, J.; Drummond, A.J. Bayesian inference of species trees from multilocus data. Mol. Biol. Evol. 2010, 27, 570–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arbogast, B.S.; Edwards, S.V.; Wakeley, J.; Beerli, P.; Slowinski, J.B. Estimating divergence times from molecular data on phylogenetic and population genetic timescales. Annu. Rev. Ecol. Syst. 2002, 33, 707–740. [Google Scholar] [CrossRef] [Green Version]
- Ogilvie, H.A.; Heled, J.; Xie, D.; Drummond, A.J. Computational performance and statistical accuracy of *BEAST and comparisons with other methods. Syst. Biol. 2016, 65, 381–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Froufe, E.; Gonçalves, D.V.; Teixeira, A.; Sousa, R.; Varandas, S.; Ghamizi, M.; Zieritz, A.; Lopes-Lima, M. Who lives where? Molecular and morphometric analyses clarify which Unio species (Unionida, Mollusca) inhabit the southwestern Palearctic. Org. Divers. Evol. 2016, 16, 597–611. [Google Scholar] [CrossRef] [Green Version]
- Bandelt, H.J.; Forster, P.; Rohl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D. popart: full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Oaks, J.R.; Linkem, C.W.; Sukumaran, J. Implications of uniformly distributed, empirically informed priors for phylogeographical model selection: A reply to Hickerson et al. Evolution 2014, 68, 3607–3617. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.; Takebayashi, N.; Qi, Y.; Hickerson, M.J. MTML-msBayes: approximate Bayesian comparative phylogeographic inference from multiple taxa and multiple loci with rate heterogeneity. Bioinformatics 2011, 12, 1. [Google Scholar] [CrossRef] [Green Version]
- Kass, R.E.; Raftery, A.E. Bayes factors. J. Am. Stat. Assoc. 1995, 90, 773–795. [Google Scholar] [CrossRef]
- Campbell, D.C.; Serb, J.M.; Buhay, J.E.; Roe, K.J.; Minton, R.L.; Lydeard, C. Phylogeny of North American amblemines (Bivalvia, Unionoida): Prodigious polyphyly proves pervasive across genera. Invertebr. Biol. 2005, 124, 131–164. [Google Scholar] [CrossRef]
- Johnson, N.A.; Smith, C.H.; Pfeiffer, J.M.; Randklev, C.R.; Williams, J.D.; Austin, J.D. Integrative taxonomy resolves taxonomic uncertainty for freshwater mussels being considered for protection under the U.S. Endangered Species Act. Sci. Rep. 2018, 8, 15892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serb, J.M.; Buhay, J.E.; Lydeard, C. Molecular systematics of the North American freshwater bivalve genus Quadrula (Unionidae: Ambleminae) based on mitochondrial ND1 sequences. Mol. Phylogenet. Evol. 2003, 28, 1–11. [Google Scholar] [CrossRef]
- King, T.L.; Eackles, M.S.; Gjetvaj, B.; Hoeh, W.R. Intraspecific phylogeography of Lasmigona subviridis (Bivalvia: Unionidae): Conservation implications of range discontinuity. Mol. Ecol. 1999, 8, S65–S78. [Google Scholar] [CrossRef]
- Thompson, J.N. The Geographic Mosaic of Coevolution; University of Chicago Press: Chicago, IL, USA, 2005; ISBN 978-0-226-79762-5. [Google Scholar]
- Hoberg, E.P. Phylogeny and historical reconstruction: Host-parasite systems as keystones in biogeography and ecology. In Biodiversity II: Understanding and Protecting our Biological Resources; John Henry Press: Washington, DC, USA, 1997; pp. 243–261. [Google Scholar]
- Inoue, K.; Lang, B.K.; Berg, D.J. Past climate change drives current genetic structure of an endangered freshwater mussel species. Mol. Ecol. 2015, 24, 1910–1926. [Google Scholar] [CrossRef]
- Beaver, C.E.; Woolnough, D.A.; Zanatta, D.T. Assessment of genetic diversity and structure among populations of Epioblasma triquetra in the Laurentian Great Lakes drainage. Freshw. Sci. 2019, 38, 527–542. [Google Scholar] [CrossRef]
- Scott, M.W.; Morris, T.J.; Zanatta, D.T. Population structure, genetic diversity, and colonization history of the eastern pondmussel, Sagittunio nasutus, in the Great Lakes drainage. Aquat. Conserv. 2020, 30, 631–646. [Google Scholar] [CrossRef]
- Whittaker, R.J.; Araújo, M.B.; Jepson, P.; Ladle, R.J.; Watson, J.E.M.; Willis, K.J. Conservation biogeography: assessment and prospect. Divers. Distrib. 2005, 11, 3–23. [Google Scholar] [CrossRef]
- Burlakova, L.E.; Karatayev, A.Y.; Karatayev, V.A.; May, M.E.; Bennett, D.L.; Cook, M.J. Biogeography and conservation of freshwater mussels (Bivalvia: Unionidae) in Texas: Patterns of diversity and threats. Divers. Distrib. 2011, 17, 393–407. [Google Scholar] [CrossRef]
- Haag, W.R. A hierarchical classification of freshwater mussel diversity in North America. J. Biogeogr. 2010, 37, 12–26. [Google Scholar] [CrossRef]
- Johnson, R.I. The systematics and zoogeography of the Unionidae (Mollusca: Bivalvia) of the southern Atlantic slope region. Harv. Univ. Mus. Comp. Zool. Bull. 1970, 140, 263–450. [Google Scholar]
- Neck, R.W. Preliminary analysis of the ecological zoogeography of the freshwater mussels of Texas. In Proceedings of the Symposium on Recent Benthological Investigations in Texas and Adjacent States; Texas Academy of Science: Austin, TX, USA, 1982; pp. 33–42. [Google Scholar]
- Sepkosk, J., Jr.; Rex, M. Distribution of freshwater mussels: Coastal rivers as biogeographic islands. Syst. Zool. 1974, 23, 165–188. [Google Scholar] [CrossRef]
- Lopes-Lima, M.; Burlakova, L.; Karatayev, A.; Gomes, A. Revisiting the North American freshwater mussel genus Quadrula sensu lato (Bivalvia Unionidae): Phylogeny, taxonomy and species delineation. Zool. Scr. 2019, 48, 313–336. [Google Scholar] [CrossRef]
- Gangloff, M.M.; Hamstead, B.A.; Abernethy, E.F.; Hartfield, P.D. Genetic distinctiveness of Ligumia recta, the black sandshell, in the Mobile River basin and implications for its conservation. Conserv. Genet. 2013, 14, 913–916. [Google Scholar] [CrossRef]
- Warren, M.L.; Burr, B.M.; Walsh, S.J.; Bart, H.L., Jr.; Cashner, R.C.; Etnier, D.A.; Freeman, B.J.; Kuhajda, B.R.; Mayden, R.L.; Robison, H.W.; et al. Diversity, distribution, and conservation status of the native freshwater fishes of the southern United States. Fisheries 2000, 25, 7–31. [Google Scholar] [CrossRef]
- Ross, S.T. The Inland Fishes of Mississippi; University of Mississippi Press: Oxford, MA, USA, 2001; ISBN 978-1-57806-246-1. [Google Scholar]
- Ennen, J.R.; Lovich, J.E.; Kreiser, B.R.; Selman, W.; Qualls, C.P. Genetic and morphological variation between populations of the Pascagoula map turtle (Graptemys gibbonsi) in the Pearl and Pascagoula rivers with description of a new species. Chelonian. Conserv. Biol. 2010, 9, 98–113. [Google Scholar] [CrossRef]
- Halas, D.; Simons, A.M. Cryptic speciation reversal in the Etheostoma zonale (Teleostei: Percidae) species group, with an examination of the effect of recombination and introgression on species tree inference. Mol. Phylogenet. Evol. 2014, 70, 13–28. [Google Scholar] [CrossRef]
- Egge, J.J.D.; Hagbo, T.J. Comparative phylogeography of Mississippi embayment fishes. PLoS ONE 2015, 10, e0116719. [Google Scholar] [CrossRef]
- Wiley, E.O.; Mayden, R.L. Species and speciation in phylogenetic systematics, with examples from the North American fish fauna. Ann. Mo. Bot. Gard. 1985, 72, 596–635. [Google Scholar] [CrossRef]
- Otvos, E.G. Coastal barriers, northern Gulf—Last Eustatic Cycle; genetic categories and development contrasts. A review. Quat. Sci. Rev. 2018, 193, 212–243. [Google Scholar] [CrossRef]
- Laporte, V.; Charlesworth, B. Effective population size and population subdivision in demographically structured populations. Genetics 2002, 162, 501–519. [Google Scholar]
- Charlesworth, B. Effective population size and patterns of molecular evolution and variation. Nat. Rev. Genet. 2009, 10, 195–205. [Google Scholar] [CrossRef]
- Moore, W.S. Inferring phylogenies from mtDNA variation: Mitochondrial-gene trees versus nuclear-gene trees. Evolution 1995, 49, 718–726. [Google Scholar] [CrossRef]
- Toews, D.P.L.; Brelsford, A. The biogeography of mitochondrial and nuclear discordance in animals. Mol. Ecol. 2012, 21, 3907–3930. [Google Scholar] [CrossRef]
- Flocks, J.; Kulp, M.; Smith, J.; Williams, S.J. Review of the geologic history of the Pontchartrain basin, northern Gulf of Mexico. J. Coast. Res. 2009, 54, 12–22. [Google Scholar] [CrossRef]
- Bolotov, I.N.; Vikhrev, I.V.; Bespalaya, Y.V.; Gofarov, M.Y.; Kondakov, A.V.; Konopleva, E.S.; Bolotov, N.N.; Lyubas, A.A. Multi-locus fossil-calibrated phylogeny, biogeography and a subgeneric revision of the Margaritiferidae (Mollusca: Bivalvia: Unionoida). Mol. Phylogenet. Evol. 2016, 103, 104–121. [Google Scholar] [CrossRef] [PubMed]
- Funk, W.C.; McKay, J.K.; Hohenlohe, P.A.; Allendorf, F.W. Harnessing genomics for delineating conservation units. Trends Ecol. Evol. 2012, 27, 489–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coates, D.J.; Byrne, M.; Moritz, C. Genetic diversity and conservation units: Dealing with the species-population continuum in the age of genomics. Front. Ecol. Evol. 2018, 6, 165. [Google Scholar] [CrossRef] [Green Version]
- Mee, J.A.; Bernatchez, L.; Reist, J.D.; Rogers, S.M.; Taylor, E.B. Identifying designatable units for intraspecific conservation prioritization: A hierarchical approach applied to the lake whitefish species complex (Coregonus spp.). Evol. Appl. 2015, 8, 423–441. [Google Scholar] [CrossRef]
- Waples, R.S. Pacific salmon, Oncorhynchus spp., and the definition of “species” under the Endangered Species Act. Mar. Fish Rev. 1991, 53, 11–22. [Google Scholar]
- Palsbøll, P.J.; Bérubé, M.; Allendorf, F.W. Identification of management units using population genetic data. Trends Ecol. Evol. 2007, 22, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Waples, R.S.; Nammack, M.; Cochrane, J.F.; Hutchings, J.A. A tale of two acts: endangered species listing practices in Canada and the United States. BioScience 2013, 63, 723–734. [Google Scholar] [CrossRef] [Green Version]
- Moritz, C. Defining ‘Evolutionarily Significant Units’ for conservation. Trends Ecol. Evol. 1994, 9, 373–375. [Google Scholar] [CrossRef]
- Grobler, P.J.; Jones, J.W.; Johnson, N.A.; Beaty, B.; Struthers, J.; Neves, R.J.; Hallerman, E.M. Patterns of genetic differentiation and conservation of the Slabside Pearlymussel, Lexingtonia dolabelloides (Lea, 1840) in the Tennessee River drainage. J. Mollus Stud. 2006, 72, 65–75. [Google Scholar] [CrossRef] [Green Version]
- McMurray, S.E.; Roe, K.J. Perspectives on the controlled propagation, augmentation, and introduction of freshwater mussels (Mollusca: Bivalvia: Unionoida). Freshw. Mollusk Biol. Conserv. 2017, 20, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Moritz, C. Strategies to protect biological diversity and the evolutionary processes that sustain it. Syst. Biol. 2002, 51, 238–254. [Google Scholar] [CrossRef]
- Neves, R.J. Propagation of endangered freshwater mussels in North America. J. Conchol. Spec. Publ. 2004, 3, 69–80. [Google Scholar]
- Snyder, N.F.R.; Derrickson, S.R.; Beissinger, S.R.; Wiley, J.W.; Smith, T.B.; Toone, W.D.; Miller, B. Limitations of captive breeding in endangered species recovery. Conserv. Biol. 1996, 10, 338–348. [Google Scholar] [CrossRef] [Green Version]
- Olden, J.D.; Kennard, M.J.; Lawler, J.J.; Poff, N.L. Challenges and opportunities in implementing managed relocation for conservation of freshwater species. Conserv. Biol. 2011, 25, 40–47. [Google Scholar] [CrossRef]
- Lopes-Lima, M.; Burlakova, L.E.; Karatayev, A.Y.; Mehler, K.; Seddon, M.; Sousa, R. Conservation of freshwater bivalves at the global scale: Diversity, threats and research needs. Hydrobiologia 2018, 810, 1–14. [Google Scholar] [CrossRef] [Green Version]



| Taxon | ID | Drainage | Source | CO1 | ND1 | ITS1 | FEM1 | UbiA |
|---|---|---|---|---|---|---|---|---|
| Potamilus fragilis | LfraAla001 | Mobile | UF438237 | MT662019 | MT669665 | MT661766 | MT669798 | MT669771 |
| Potamilus fragilis | LfraAmi040 | Pontchartrain | UF439330 | MT662020 | MT669666 | MT661773 | MT669778 | MT669751 |
| Potamilus fragilis | LfraAmi041 | Pontchartrain | UF439352 | MT662021 | MT669667 | |||
| Potamilus fragilis | LfraAmi042 | Pontchartrain | UF439352 | MT662022 | MT669668 | |||
| Potamilus fragilis | LfraPrl043 | Pearl | UF439332 | MT662023 | MT669669 | |||
| Potamilus fragilis | LfraPrl044 | Pearl | UF439332 | MT662024 | MT669670 | MT661780 | MT669785 | MT669758 |
| Potamilus fragilis | LfraPrl045 | Pearl | UF439365 | MT662025 | MT669671 | |||
| Potamilus fragilis | LfraPrl046 | Pearl | UF439343 | MT662026 | MT669672 | |||
| Potamilus fragilis | LfraPrl047 | Pearl | UF439343 | MT662027 | MT669673 | |||
| Potamilus fragilis | LfraPrl048 | Pearl | UF439343 | MT662028 | MT669674 | |||
| Potamilus fragilis | LfraAmi057 | Pontchartrain | UF439529 | MT662029 | MT669675 | |||
| Potamilus fragilis | LfraAmi058 | Pontchartrain | UF439529 | MT662030 | MT669676 | |||
| Potamilus fragilis | LfraAmi059 | Pontchartrain | UF439529 | MT662031 | MT669677 | |||
| Potamilus fragilis | LfraMob063 | Mobile | UF439528 | MT662033 | MT669679 | |||
| Potamilus fragilis | LfraMob064 | Mobile | UF439528 | MT662032 | MT669678 | |||
| Potamilus fragilis | LfraMob065 | Mobile | Uncatologed | MT662034 | MT669680 | MT661792 | MT669797 | MT669770 |
| Potamilus inflatus | PinfMob001 | Mobile | UF439131 | MT662002 | MT669647 | MT661768 | MT669773 | MT669746 |
| Potamilus inflatus | PinfMob002 | Mobile | UF439131 | MK044952 | MK045103 | MK036203 | MT669774 | MT669747 |
| Potamilus inflatus | PinfMob003 | Mobile | UF439131 | MT662003 | MT669648 | MT661769 | MT669775 | MT669748 |
| Potamilus inflatus | PinfMob004 | Mobile | UF439131 | MK044953 | MK045104 | MK036204 | SRR10579071 | SRR10579071 |
| Potamilus inflatus | PinfMob005 | Mobile | UF439131 | MT662004 | MT669649 | MT661770 | MT669776 | MT669749 |
| Potamilus inflatus | PinfMob006 | Mobile | UF439131 | MT662005 | MT669650 | MT661771 | MT669777 | MT669750 |
| Potamilus inflatus | PinfAmi010 | Pontchartrain | UF439530 | MT662006 | MT669651 | MT661774 | MT669779 | MT669752 |
| Potamilus inflatus | PinfAmi011 | Pontchartrain | UF439530 | MT662007 | MT669652 | MT661775 | MT669780 | MT669753 |
| Potamilus inflatus | PinfAmi012 | Pontchartrain | UF439531 | MT662008 | MT669653 | MT661776 | MT669781 | MT669754 |
| Potamilus inflatus | PinfAmi013 | Pontchartrain | UF439532 | MT662009 | MT669654 | MT661777 | MT669782 | MT669755 |
| Potamilus inflatus | PinfAmi014 | Pontchartrain | UF439532 | MT662010 | MT669655 | MT661778 | MT669783 | MT669756 |
| Potamilus inflatus | PinfAmi015 | Pontchartrain | UF439533 | MT662011 | MT669656 | MT661779 | MT669784 | MT669757 |
| Potamilus inflatus | PinfMob019 | Mobile | UF439514 | MT662012 | MT669657 | MT661783 | MT669788 | MT669761 |
| Potamilus inflatus | PinfMob020 | Mobile | UF439514 | MT662013 | MT669658 | MT661784 | MT669789 | MT669762 |
| Potamilus inflatus | PinfMob021 | Mobile | UF439514 | MT662014 | MT669659 | MT661785 | MT669790 | MT669763 |
| Potamilus inflatus | PinfMob022 | Mobile | UF439514 | MT662015 | MT669660 | MT661786 | MT669791 | MT669764 |
| Potamilus inflatus | PinfMob023 | Mobile | UF439514 | MT662016 | MT669661 | MT661787 | MT669792 | MT669765 |
| Potamilus inflatus | PinfMob017 | Mobile | UF439513 | MT662017 | MT669662 | MT661788 | MT669793 | MT669766 |
| Potamilus inflatus | PinfMob018 | Mobile | UF439513 | MT662018 | MT669663 | MT661789 | MT669794 | MT669767 |
| Potamilus inflatus | PinfMob016 | Mobile | UA2696 | MT669664 | MT661781 | MT669786 | MT669759 | |
| Potamilus purpuratus | PpurPas001 | Pascagoula | UF438434 | MT662035 | MT669681 | |||
| Potamilus purpuratus | PpurPrl022 | Pearl | UF439145 | MT662036 | MT669682 | |||
| Potamilus purpuratus | PpurPrl023 | Pearl | UF439145 | MK044960 | MK045111 | MK036211 | MT669799 | MT669772 |
| Potamilus purpuratus | PpurPrl024 | Pearl | UF439145 | MK044961 | MK045112 | MK036212 | ||
| Potamilus purpuratus | PpurPrl025 | Pearl | UF439145 | MT662037 | MT669683 | |||
| Potamilus purpuratus | PpurPrl026 | Pearl | UF439145 | MT662038 | MT669684 | MT661767 | ||
| Potamilus purpuratus | PpurAmi038 | Pontchartrain | UF439452 | MT662039 | MT669685 | |||
| Potamilus purpuratus | PpurAmi039 | Pontchartrain | UF439452 | MT662040 | MT669686 | |||
| Potamilus purpuratus | PpurAmi040 | Pontchartrain | UF439452 | MT662041 | MT669687 | |||
| Potamilus purpuratus | PpurAmi041 | Pontchartrain | UF439452 | MT662042 | MT669688 | |||
| Potamilus purpuratus | PpurAmi042 | Pontchartrain | UF439452 | MT662043 | MT669689 | |||
| Potamilus purpuratus | PpurAmi043 | Pontchartrain | UF439453 | MT662044 | MT669690 | |||
| Potamilus purpuratus | PpurAmi044 | Pontchartrain | UF439453 | MT662045 | MT669691 | |||
| Potamilus purpuratus | PpurAmi045 | Pontchartrain | UF439453 | MT662046 | MT669692 | MT661772 | SRR10579081 | SRR10579081 |
| Potamilus purpuratus | PpurAmi046 | Pontchartrain | UF439453 | MT662047 | MT669693 | |||
| Potamilus purpuratus | PpurAmi047 | Pontchartrain | UF439453 | MT662048 | MT669694 | |||
| Potamilus purpuratus | PpurAmi048 | Pontchartrain | UF439454 | MT662049 | MT669695 | |||
| Potamilus purpuratus | PpurAmi049 | Pontchartrain | UF439454 | MT662050 | MT669696 | |||
| Potamilus purpuratus | PpurAmi050 | Pontchartrain | UF439454 | MT662051 | MT669697 | |||
| Potamilus purpuratus | PpurAmi051 | Pontchartrain | UF439454 | MT662052 | MT669698 | |||
| Potamilus purpuratus | PpurPrl052 | Pearl | UF439456 | MT662053 | MT669699 | |||
| Potamilus purpuratus | PpurPrl053 | Pearl | UF439456 | MT662054 | MT669700 | |||
| Potamilus purpuratus | PpurPrl054 | Pearl | UF439457 | MT662055 | MT669701 | |||
| Potamilus purpuratus | PpurPrl055 | Pearl | UF439457 | MT662056 | MT669702 | |||
| Potamilus purpuratus | PpurPrl056 | Pearl | UF439457 | MT662057 | MT669703 | |||
| Potamilus purpuratus | PpurPrl057 | Pearl | UF439457 | MT662058 | MT669704 | |||
| Potamilus purpuratus | PpurPrl058 | Pearl | UF439457 | MT662059 | MT669705 | |||
| Potamilus purpuratus | PpurPrl059 | Pearl | UF439456 | MT662060 | MT669706 | |||
| Potamilus purpuratus | PpurPrl060 | Pearl | UF439456 | MT662061 | MT669707 | |||
| Potamilus purpuratus | PpurPrl061 | Pearl | UF439456 | MT662062 | MT669708 | |||
| Potamilus purpuratus | PpurPrl062 | Pearl | UF439456 | MT662063 | MT669709 | |||
| Potamilus purpuratus | PpurPrl063 | Pearl | UF439456 | MT662064 | MT669710 | |||
| Potamilus purpuratus | PpurPrl064 | Pearl | UF439458 | MT662065 | MT669711 | |||
| Potamilus purpuratus | PpurPrl065 | Pearl | UF439459 | MT662066 | MT669712 | |||
| Potamilus purpuratus | PpurPrl066 | Pearl | UF439459 | MT662067 | MT669713 | |||
| Potamilus purpuratus | PpurPrl067 | Pearl | UF439459 | MT662068 | MT669714 | |||
| Potamilus purpuratus | PpurPrl068 | Pearl | UF439459 | MT662069 | MT669715 | |||
| Potamilus purpuratus | PpurPrl069 | Pearl | UF439459 | MT662070 | MT669716 | |||
| Potamilus purpuratus | PpurMob081 | Mobile | UA62 | MT662071 | MT669717 | |||
| Potamilus purpuratus | PpurMob082 | Mobile | UA2469 | MT662072 | MT669718 | |||
| Potamilus purpuratus | PpurMob083 | Mobile | UA2510 | MT662073 | MT669719 | |||
| Potamilus purpuratus | PpurMob084 | Mobile | UA2562 | MT662074 | MT669720 | |||
| Potamilus purpuratus | PpurMob085 | Mobile | UA2740 | MT662075 | MT669721 | MT661782 | MT669787 | MT669760 |
| Potamilus purpuratus | PpurMob086 | Mobile | UA3100 | MT662076 | MT669722 | |||
| Potamilus purpuratus | PpurMob087 | Mobile | UA3123 | MT662077 | MT669723 | |||
| Potamilus purpuratus | PpurMob088 | Mobile | UA3205 | MT662078 | MT669724 | |||
| Potamilus purpuratus | PpurMob089 | Mobile | UA3417 | MT662079 | MT669725 | |||
| Potamilus purpuratus | PpurMob090 | Mobile | UA3482 | MT662080 | MT669726 | |||
| Potamilus purpuratus | PpurPas097 | Pascagoula | UF439510 | MT662081 | MT669727 | |||
| Potamilus purpuratus | PpurPas098 | Pascagoula | UF439510 | MT662082 | MT669728 | |||
| Potamilus purpuratus | PpurPas099 | Pascagoula | UF439510 | MT662083 | MT669729 | |||
| Potamilus purpuratus | PpurPas100 | Pascagoula | UF439510 | MT662084 | MT669730 | |||
| Potamilus purpuratus | PpurPas101 | Pascagoula | UF439510 | MT662085 | MT669731 | MT661790 | MT669795 | MT669768 |
| Potamilus purpuratus | PpurPas102 | Pascagoula | UF439510 | MT662086 | MT669732 | |||
| Potamilus purpuratus | PpurPas103 | Pascagoula | UF439510 | MT662087 | MT669733 | |||
| Potamilus purpuratus | PpurMob107 | Mobile | UF439527 | MT662088 | MT669734 | MT661791 | MT669796 | MT669769 |
| Potamilus purpuratus | PpurMob108 | Mobile | UF439527 | MT662089 | MT669735 | |||
| Potamilus purpuratus | PpurMob109 | Mobile | UF439527 | MT662090 | MT669736 | |||
| Potamilus purpuratus | PpurMob110 | Mobile | UF439527 | MT662091 | MT669737 | |||
| Potamilus purpuratus | PpurMob111 | Mobile | UF439527 | MT662092 | MT669738 | |||
| Potamilus purpuratus | PpurMob112 | Mobile | UF439527 | MT662093 | MT669739 | |||
| Potamilus purpuratus | PpurMob113 | Mobile | UF439527 | MT662094 | MT669740 | |||
| Potamilus purpuratus | PpurMob114 | Mobile | UF439527 | MT662095 | MT669741 | |||
| Potamilus purpuratus | PpurMob115 | Mobile | UF439527 | MT662096 | MT669742 | |||
| Potamilus purpuratus | PpurMob116 | Mobile | UF439527 | MT662097 | MT669743 | |||
| Potamilus purpuratus | PpurMob117 | Mobile | UF439527 | MT662098 | MT669744 | |||
| Potamilus purpuratus | PpurMob118 | Mobile | UF439527 | MT662099 | MT669745 |
| Locus | Primers | Source | Conditions |
|---|---|---|---|
| CO1 | F: 5′-GTTCCACAAATCATAAGGATATTGG-3′ | [68] | [69] |
| R: 5′-TACACCTCAGGGTGACCAAAAAACCA-3′ | |||
| ND1 | F: 5′-TGGCAGAAAAGTGCATCAGATTAAAGC-3′ | [70] | [70] |
| R: 5′-CCTGCTTGGAAGGCAAGTGTACT-3′ | |||
| ITS1 | F: 5′-AAAAAGCTTCCGTAGGTGAACCTGCG-3′ | [71] | [71] |
| R: 5′-AGCTTGCTGCGTTCTTCATCG-3′ | |||
| FEM1 | F: 5′- GTRATGGAGTATCGCAGTGT-3′ | [44] | [44] |
| R: 5′-ACRCTYTTCCTGTTAACATC-3′ | |||
| UbiA | F: 5′- TTTACTCCTGTTGCACTTGGGA-3′ | [44] | [44] |
| R: 5′-AGCATCTGTCATGAAGACTCCAAC-3′ |
| Taxon | N Mobile | N PPP | AMOVA between | AMOVA within | Distance between (Uncorrected p) |
|---|---|---|---|---|---|
| Potamilus fragilis | 4 | 12 | 80.9% | 19.1% | 1.11% |
| Potamilus inflatus | 13 | 6 | 98.9% | 1.1% | 2.33% |
| Potamilus purpuratus | 22 | 45 | 96.3% | 3.7% | 1.31% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith, C.H.; Johnson, N.A. A Comparative Phylogeographic Approach to Facilitate Recovery of an Imperiled Freshwater Mussel (Bivalvia: Unionida: Potamilus inflatus). Diversity 2020, 12, 281. https://doi.org/10.3390/d12070281
Smith CH, Johnson NA. A Comparative Phylogeographic Approach to Facilitate Recovery of an Imperiled Freshwater Mussel (Bivalvia: Unionida: Potamilus inflatus). Diversity. 2020; 12(7):281. https://doi.org/10.3390/d12070281
Chicago/Turabian StyleSmith, Chase H., and Nathan A. Johnson. 2020. "A Comparative Phylogeographic Approach to Facilitate Recovery of an Imperiled Freshwater Mussel (Bivalvia: Unionida: Potamilus inflatus)" Diversity 12, no. 7: 281. https://doi.org/10.3390/d12070281
APA StyleSmith, C. H., & Johnson, N. A. (2020). A Comparative Phylogeographic Approach to Facilitate Recovery of an Imperiled Freshwater Mussel (Bivalvia: Unionida: Potamilus inflatus). Diversity, 12(7), 281. https://doi.org/10.3390/d12070281