Phenotypic Variation in Endangered Texas Salamanders: Application of Model-Based Clustering for Identifying Species and Hybrids
Abstract
:1. Introduction
2. Materials and Methods
2.1. Photographic Dataset and Landmark Digitization
2.2. Assignment of Individuals to Species/Groups and Assessment of Phenotypic Variation
2.3. Microgeographic Variation in Eurycea Sosorum
3. Results
3.1. Model-Based Clustering
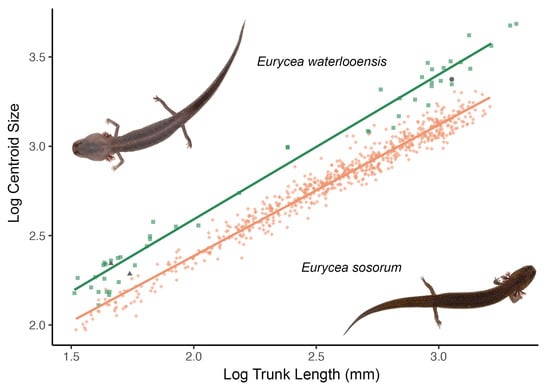
3.2. Allometry and Phenotypic Variation
3.3. Microgeographic Variation in E. sosorum
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bensmail, H.; Celeux, G.; Raftery, A.E.; Robert, C.P. Inference in model-based cluster analysis. Stat. Comput. 1997, 7, 1–10. [Google Scholar] [CrossRef]
- Bing, X.; Bunea, F.; Royer, M.; Das, J. Latent model-based clustering for biological discovery. iScience 2019, 14, 125–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, W.; Lin, J.; Le, C.T. Model-based cluster analysis of microarray gene-expression data. Genome Biol. 2002, 3, research0009.1. [Google Scholar]
- Medvedovic, M.; Yeung, K.Y.; Bumgarner, R.E. Bayesian mixture model-based clustering of replicated microarray data. Bioinformatics 2004, 20, 1222–1232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ezard, T.H.G.; Pearson, P.N.; Purvis, A. Algorithmic approaches to aid species’ delimitation in multidimensional morphospace. BMC Evol. Biol. 2010, 10, 175. [Google Scholar] [CrossRef] [Green Version]
- Meik, J.M.; Streicher, J.W.; Lawing, A.M.; Flores-Villela, O.; Fujita, M.K. Limitations of climatic data for inferring species boundaries: Insights from speckled rattlesnakes. PLoS ONE 2015, 10, e0131435. [Google Scholar] [CrossRef] [Green Version]
- Cadena, C.D.; Zapata, F.; Jiménez, I. Issues and perspectives in species delimitation using phenotypic data: Atlantean evolution in Darwin’s finches. Syst. Biol. 2018, 67, 181–194. [Google Scholar] [CrossRef]
- Fraley, C.; Raftery, A.E. Model-based clustering, discriminant analysis, and density estimation. J. Am. Stat. Assoc. 2002, 97, 611–631. [Google Scholar] [CrossRef]
- Scrucca, L.; Fop, M.; Murphy, T.B.; Raftery, A.E. mclust 5: Clustering, classification and density estimation using Gaussian finite mixture models. R J. 2016, 8, 289–317. [Google Scholar] [CrossRef] [Green Version]
- Ronan, T.; Qi, Z.; Naegle, K.M. Avoiding common pitfalls when clustering biological data. Sci. Signal. 2016, 9, re6. [Google Scholar] [CrossRef] [Green Version]
- Chippindale, P.T.; Price, A.H.; Wiens, J.J.; Hillis, D.M. Phylogenetic relationships and systematic revision of central Texas hemidactyliine plethodontid salamanders. Herpetol. Monogr. 2000, 14, 1–80. [Google Scholar] [CrossRef] [Green Version]
- Bendik, N.F.; Meik, J.M.; Gluesenkamp, A.G.; Roelke, C.E.; Chippindale, P.T. Biogeography, phylogeny, and morphological evolution of central Texas cave and spring salamanders. BMC Evol. Biol. 2013, 13, 201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devitt, T.J.; Wright, A.M.; Cannatella, D.C.; Hillis, D.M. Species delimitation in endangered groundwater salamanders: Implications for aquifer management and biodiversity conservation. Proc. Natl. Acad. Sci. USA 2019, 116, 2624–2633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- City of Austin. 2018 Annual Report U.S. Fish and Wildlife Service Scientific Permit (TE-833851); Watershed Protection Department: Austin, TX, USA, 2018. Available online: www.austintexas.gov/edims/pio/document.cfm?id=314140 (accessed on 20 June 2020).
- Hillis, D.M.; Chamberlain, D.A.; Wilcox, T.P.; Chippindale, P.T. A New species of subterranean blind salamander (Plethodontidae: Hemidactyliini: Eurycea: Typhlomolge) from Austin, Texas, and a systematic revision of central Texas paedomorphic salamanders. Herpetologica 2001, 57, 266–280. [Google Scholar]
- Wiens, J.J.; Chippindale, P.T.; Hillis, D.M. When are phylogenetic analyses misled by convergence? A case study in Texas cave salamanders. Syst. Biol. 2003, 52, 501–514. [Google Scholar] [CrossRef]
- Chippindale, P.T.; Price, A.H.; Hillis, D.M. A new species of perennibranchiate salamander (Eurycea: Plethodontidae) from Austin, Texas. Herpetologica 1993, 49, 248–259. [Google Scholar]
- Bendik, N.F.; Morrison, T.A.; Gluesenkamp, A.G.; Sanders, M.S.; O’Donnell, L.J. Computer-assisted photo identification outperforms visible implant elastomers in an endangered salamander, Eurycea tonkawae. PLoS ONE 2013, 8, e59424. [Google Scholar] [CrossRef] [Green Version]
- Bendik, N.F.; Dries, L.A. Density-dependent and density-independent drivers of population change in Barton Springs salamanders. Ecol. Evol. 2018, 8, 5912–5923. [Google Scholar] [CrossRef]
- Bendik, N.F.; Chamberlain, D.A.; Devitt, T.J.; Robinson, D. Barton Springs and Austin Blind Salamander Capture-Recapture Study: Interim Report Submitted to City of Austin Watershed Protection Department. 2019. Available online: http://www.austintexas.gov/watershed_protection/publications/document.cfm?id=339918 (accessed on 20 June 2020).
- Rohlf, F.J. TpsDig2. 2010. Available online: https://tpsdig2.software.informer.com/download (accessed on 20 June 2020).
- Cardini, A. Lost in the other half: Improving accuracy in geometric morphometric analyses of one side of bilaterally symmetric structures. Syst. Biol. 2016, 65, 1096–1106. [Google Scholar] [CrossRef]
- Lawing, A.M.; Polly, P.D. Geometric morphometrics: Recent applications to the study of evolution and development. J. Zool. 2010, 280, 1–7. [Google Scholar] [CrossRef]
- Kaliontzopoulou, A. Geometric morphometrics in herpetology: Modern tools for enhancing the study of morphological variation in amphibians and reptiles. Basic Appl. Herpetol. 2011, 25, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Zelditch, M.L.; Swiderski, D.L.; Sheets, H.D. Geometric Morphometrics for Biologists: A Primer, 2nd ed.; Academic Press: San Diego, CA, USA, 2012; pp. 105–128. [Google Scholar]
- Björklund, M. Be careful with your principal components. Evolution 2019, 73, 2151–2158. [Google Scholar] [CrossRef] [PubMed]
- Rieseberg, L.H.; Archer, M.A.; Wayne, R.K. Transgressive segregation, adaptation and speciation. Heredity 1999, 83, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Bell, M.A.; Travis, M.P. Hybridization, transgressive segregation, genetic covariation, and adaptive radiation. Trends Ecol. Evol. 2005, 20, 358–361. [Google Scholar] [CrossRef] [PubMed]
- Bruce, P.; Bruce, A. Practical Statistics for Data Scientists: 50 Essential Concepts, 1st ed.; O’Reilly Media Inc.: Sebastopol, CA, USA, 2017; pp. 270–276. [Google Scholar]
- Fruhwirth-Schnatter, S.; Celeux, G.; Robert, C.P. Model-based clustering. In Handbook of Mixture Analysis; Chapman and Hall: New York, NY, USA, 2019; pp. 158–184. [Google Scholar]
- R: A Language and Environment for Statistical Computing. Available online: https://www.gbif.org/tool/81287/r-a-language-and-environment-for-statistical-computing (accessed on 20 June 2020).
- Shapes: Statistical Shape Analysis. Available online: https://cran.r-project.org/package=shapes (accessed on 20 June 2020).
- Fox, J.; Friendly, M.; Monette, G. Visualizing hypothesis tests in multivariate linear models: The heplots package for R. Comput. Stat. 2008, 24, 233–246. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, R.W.; Reddell, J.R. Eurycea tridentifera, a new species of troglobitic salamander from Texas and a reclassification of Typhlomolge rathbuni. Tex. J. Sci. 1965, 17, 12–27. [Google Scholar]
- Mitchell, R.W.; Smith, R.E. Some aspects of the osteology and evolution of the neotenic spring and cave salamanders (Eurycea, Plethodontidae) of central Texas. Tex. J. Sci. 1972, 23, 343–362. [Google Scholar]
- Sweet, S.S. Secondary contact and hybridization in the Texas cave salamanders Eurycea neotenes and E. tridentifera. Copeia 1984, 1984, 428–441. [Google Scholar] [CrossRef] [Green Version]
- Coyne, J.A.; Orr, H.A. Speciation; Sinauer Assoc.: Sunderland, MA, USA, 2004; pp. 1–25. [Google Scholar]
- Nichols, P.; Genner, M.J.; Oosterhout, C.V.; Smith, A.; Parsons, P.; Sungani, H.; Swanstrom, J.; Joyce, D.A. Secondary contact seeds phenotypic novelty in cichlid fishes. Proc. R. Soc. B. 2015, 282, 20142272. [Google Scholar] [CrossRef] [Green Version]
- McDermid, K.; Sprouse, P.; Krejca, J. Geographical distribution: Eurycea sosorum (Barton Springs salamander). Herpetol. Rev. 2015, 556–578. [Google Scholar]
- Devitt, T.; Nissen, B.D. New occurrence records for Eurycea sosorum Chippindale, Price & Hillis, 1993 (Caudata, Plethodontidae) in Travis and Hays counties, Texas, USA. Check List 2018, 14, 297–301. [Google Scholar]
- Santos, C.M.D.; Amorim, D.S.; Klassa, B.; Fachin, D.A.; Nihei, S.S.; De Carvalho, C.J.B.; Falaschi, R.L.; Mello-Patiu, C.A.; Couri, M.S.; Iliveira, S.S.; et al. On typeless species and the perils of fast taxonomy. Syst. Entomol. 2016, 41, 511–515. [Google Scholar] [CrossRef] [Green Version]
- Porto, A.; Voje, K.L. ML-morph: A fast, accurate and general approach for automated detection and landmarking of biological structures in images. Methods Ecol. Evol. 2020, 11, 500–512. [Google Scholar] [CrossRef] [Green Version]






| Source | η2 | Group df | Pillai | ~F-Value | p-Value |
|---|---|---|---|---|---|
| Season | 0.04 | 3 | 0.12 | 4.97 | <0.001 |
| Locality | 0.15 | 3 | 0.45 | 20.40 | <0.001 |
| Size | 0.49 | 1 | 0.49 | 110.97 | <0.001 |
| Locality:Size | 0.02 | 3 | 0.07 | 2.72 | <0.001 |
| Season:Size | 0.01 | 3 | 0.04 | 1.37 | 0.14 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Strom, D.M.; Bendik, N.F.; Chamberlain, D.A.; Watson, J.A.; Meik, J.M. Phenotypic Variation in Endangered Texas Salamanders: Application of Model-Based Clustering for Identifying Species and Hybrids. Diversity 2020, 12, 297. https://doi.org/10.3390/d12080297
Strom DM, Bendik NF, Chamberlain DA, Watson JA, Meik JM. Phenotypic Variation in Endangered Texas Salamanders: Application of Model-Based Clustering for Identifying Species and Hybrids. Diversity. 2020; 12(8):297. https://doi.org/10.3390/d12080297
Chicago/Turabian StyleStrom, Donella M., Nathan F. Bendik, Dee Ann Chamberlain, Jessica A. Watson, and Jesse M. Meik. 2020. "Phenotypic Variation in Endangered Texas Salamanders: Application of Model-Based Clustering for Identifying Species and Hybrids" Diversity 12, no. 8: 297. https://doi.org/10.3390/d12080297
APA StyleStrom, D. M., Bendik, N. F., Chamberlain, D. A., Watson, J. A., & Meik, J. M. (2020). Phenotypic Variation in Endangered Texas Salamanders: Application of Model-Based Clustering for Identifying Species and Hybrids. Diversity, 12(8), 297. https://doi.org/10.3390/d12080297