Recognition and Characterization of Forest Plant Communities through Remote-Sensing NDVI Time Series
Abstract
:1. Introduction
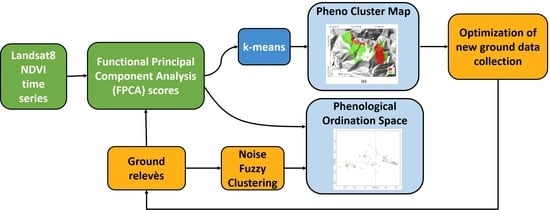
2. Materials and Methods
2.1. Study Area
2.2. Remotely Sensed NDVI Times Series
2.3. NDVI Seasonality: Functional Principal Component Analysis of NDVI Time Series
2.4. Phytosociological Data Collection
2.4.1. Data-Driven Ground Data Collection
2.4.2. Ground Dataset
2.5. Plant Communities: Ground Data Classification
2.6. NDVI Seasonality and Forest Plant Community Relationships
3. Results
3.1. Phytosociological Data Collection
3.2. Forest Plant Communities and NDVI Seasonality Relationships
4. Discussion
4.1. General Discussion
4.2. Comparison of Mount Conero and Mount Valmontagnana Forest Plant Communities
5. Conclusions and Future Works
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Plant Communities (Cluster) | 6 | 4 | 1 | 3 | 2 | 5 |
|---|---|---|---|---|---|---|
| N. relevés | 10 | 7 | 7 | 6 | 16 | 18 |
| Mean Richness | 30 | 34 | 27 | 28 | 17 | 13 |
| H trees (m) | 13.8 | 9.7 | 7 | 12 | 12 | 8.8 |
| Ostrya carpinifolia | 100 | 100 | 86 | 100 | 100 | |
| Quercus ilex | 60 | 100 | 100 | 100 | 100 | 100 |
| Species of the class Quercetea ilicis | ||||||
| Laurus nobilis | 17 | 41 | 5 | |||
| Phillyrea latifolia | 29 | 57 | 6 | 47 | ||
| Pistacia lentiscus | 47 | |||||
| Arbutus unedo | 53 | 95 | ||||
| Pistacia terebinthus ssp. terebinthus | 30 | 14 | 5 | |||
| Smilax aspera | 14 | 29 | 83 | 100 | 100 | |
| Rosa sempervirens | 67 | 29 | 47 | |||
| Viburnum tinus ssp. tinus | 14 | 17 | 94 | 100 | ||
| Rubia peregrina | 40 | 57 | 100 | 100 | 82 | 74 |
| Cephalanthera longifolia | 10 | 33 | 6 | 11 | ||
| Asparagus acutifolius | 100 | 71 | 100 | 100 | 88 | 100 |
| Lonicera implexa | 16 | |||||
| Teucrium flavum | 14 | 11 | ||||
| Pinus halepensis | 11 | |||||
| Cephalanthera rubra | 6 | |||||
| Clematis flammula | 5 | |||||
| Species of the class Querco-Fagetea | ||||||
| Euonymus latifolius | 30 | |||||
| Staphylea pinnata | 30 | |||||
| Quercus cerris | 100 | 29 | 14 | |||
| Sorbus aria | 20 | 71 | 29 | |||
| Acer campestre | 10 | 71 | 14 | 17 | ||
| Acer monspessulanum ssp. monspessulanum | 50 | 71 | 100 | |||
| Euonymus europaeus | 20 | 33 | ||||
| Prunus avium | 33 | |||||
| Ilex aquifolium | 20 | 14 | 47 | |||
| Acer opalus ssp. obtusatum | 90 | 86 | 29 | 100 | 65 | 5 |
| Quercus pubescens | 50 | 100 | 71 | 100 | 53 | 42 |
| Corylus avellana | 30 | 14 | ||||
| Fraxinus ornus ssp. ornus | 100 | 100 | 100 | 100 | 100 | 100 |
| Sorbus torminalis | 50 | 29 | 17 | 35 | 5 | |
| Sorbus domestica | 20 | 14 | 50 | 18 | 16 | |
| Dioscorea communis | 50 | 14 | 71 | |||
| Hedera helix ssp. helix | 90 | 57 | 86 | 100 | 100 | 42 |
| Clematis vitalba | 20 | 29 | 17 | |||
| Cornus mas | 100 | 43 | 43 | 17 | ||
| Rosa arvensis | 20 | 29 | ||||
| Lonicera xylosteum | 50 | 86 | 29 | |||
| Lonicera caprifolium | 20 | 29 | ||||
| Rubus hirtus | 20 | 29 | ||||
| Prunus spinosa ssp. spinosa | 20 | 71 | ||||
| Lonicera etrusca | 10 | 29 | 14 | 100 | 6 | 5 |
| Lathyrus venetus | 40 | 14 | 29 | |||
| Helleborus viridis ssp. bocconei | 90 | 43 | 14 | |||
| Lilium bulbiferum ssp. croceum | 20 | 43 | ||||
| Hieracium cf. racemosum | 43 | |||||
| Stachys officinalis | 10 | 14 | 43 | 67 | ||
| Brachypodium sylvaticum | 14 | 83 | ||||
| Buglossoides purpurocaerulea | 30 | 14 | 14 | 67 | ||
| Campanula trachelium ssp. trachelium | 10 | 33 | ||||
| Ruscus hypoglossum | 17 | 65 | 5 | |||
| Helleborus foetidus ssp. foetidus | 40 | 43 | 14 | |||
| Primula vulgaris | 50 | 43 | 14 | 50 | ||
| Melica uniflora | 60 | 29 | 86 | |||
| Euphorbia amygdaloides | 70 | 71 | 14 | 12 | ||
| Viola reichenbachiana | 60 | 29 | 14 | 67 | 6 | |
| Hepatica nobilis | 90 | 57 | 43 | 50 | 12 | |
| Viola alba ssp. dehnhardtii | 80 | 100 | 86 | 100 | 29 | 5 |
| Melittis melissophyllum | 80 | 14 | 86 | 83 | 12 | |
| Mercurialis perennis | 30 | 29 | 14 | 41 | ||
| Cyclamen hederifolium | 70 | 29 | 86 | 50 | 41 | 21 |
| Cyclamen repandum S. et S. | 43 | 17 | 65 | |||
| Daphne laureola | 80 | 100 | 29 | 100 | 53 | 16 |
| Sanicula europaea | 30 | 14 | 17 | |||
| Mycelis muralis ssp. muralis | 14 | 17 | ||||
| Fagus sylvatica ssp. sylvatica | 10 | 14 | ||||
| Stellaria holostea ssp. holostea | 14 | |||||
| Crataegus laevigata | 10 | |||||
| Doronicum columnae | 14 | |||||
| Euphorbia dulcis | 10 | |||||
| Fragaria vesca L. | 17 | |||||
| Geum urbanum | 14 | |||||
| Hieracium murorum | 14 | |||||
| Laburnum anagyroides | 14 | 14 | ||||
| Rubus caesius | 14 | |||||
| Tilia cordata | 10 | |||||
| Species of the class Rhamno-Prunetea | ||||||
| Rosa canina | 29 | 14 | ||||
| Juniperus communis | 20 | 29 | ||||
| Cytisophyllum sessilifolium | 10 | 57 | 17 | |||
| Juniperus oxycedrus | 30 | 86 | 43 | 37 | ||
| Cotinus coggygria | 60 | 57 | 43 | 5 | ||
| Crataegus monogyna | 80 | 57 | 43 | 67 | 12 | |
| Rubus ulmifolius | 14 | 50 | 6 | |||
| Cornus sanguinea | 14 | 14 | 83 | 12 | ||
| Coronilla emerus ssp. Emeroides | 50 | 57 | 100 | 100 | 47 | 79 |
| Ligustrum vulgare | 43 | |||||
| Amelanchier ovalis | 14 | |||||
| Colutea arborescens | 14 | |||||
| Rhamnus cathartica | 14 | |||||
| Rhamnus fallax | 14 | |||||
| Spartium junceum | 5 | |||||
| Other species | ||||||
| Pteridium aquilinum ssp. aquilinum | 29 | |||||
| Echinops siculus | 43 | |||||
| Peucedanum oreoselinum | 10 | 57 | ||||
| Asplenium trichomanes | 43 | |||||
| Asplenium onopteris | 30 | 14 | 100 | 17 | 6 | 5 |
| Ceterach officinarum | 14 | 86 | 5 | |||
| Solidago virgaurea | 10 | 33 | ||||
| Digitalis micrantha | 20 | 43 | 29 | |||
| Brachypodium rupestre | 40 | 86 | 29 | 67 | 5 | |
| Polypodium vulgare | 20 | 29 | 29 | |||
| Arabis turrita | 43 | 43 | 5 | |||
| Osyris alba | 29 | 29 | 12 | 63 | ||
| Epipactis helleborine | 29 | 33 | 12 | 11 | ||
| Ruscus aculeatus | 90 | 86 | 100 | 83 | 100 | 95 |
| Plant Communities (Cluster) | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| 1 | - | - | - | - | - | - |
| 2 | 0.0015 | - | - | - | - | - |
| 3 | 0.0056 | 0.0015 | - | - | - | - |
| 4 | 0.0021 | 0.0015 | 0.0020 | - | - | - |
| 5 | 0.0015 | 0.0015 | 0.0015 | 0.0015 | - | - |
| 6 | 0.0015 | 0.0015 | 0.0015 | 0.0074 | 0.0015 | - |
References
- Braun-Blanquet, J.; Conard, H.S.; Fuller, G.D. Plant Sociology, the Study of Plant Communities, 1st ed.; McGraw-Hill Book Company, Inc.: New York, NY, USA, 1932. [Google Scholar]
- Biondi, E.; Blasi, C.; Allegrezza, M.; Anzellotti, I.; Azzella, M.M.; Carli, E.; Casavecchia, S.; Copiz, R.; Del Vico, E.; Facioni, L.; et al. Plant communities of Italy: The Vegetation Prodrome. Plant Biosyst. 2014, 148, 728–814. [Google Scholar] [CrossRef] [Green Version]
- Mucina, L.; Bültmann, H.; Dierßen, K.; Theurillat, J.P.; Raus, T.; Čarni, A.; Šumberová, K.; Willner, W.; Dengler, J.; García, R.G.; et al. Vegetation of Europe: Hierarchical floristic classification system of vascular plant, bryophyte, lichen, and algal communities. Appl. Veg. Sci. 2016, 19, 3–264. [Google Scholar] [CrossRef]
- Biondi, E. Phytosociology today: Methodological and conceptual evo lution. Plant Biosyst. 2011, 145, 19–29. [Google Scholar] [CrossRef]
- Rivas-Martínez, S. Notions on dynamic-catenal phytosociology as a basis of landscape science. Plant Biosyst. 2005, 139, 135–144. [Google Scholar] [CrossRef]
- Chytrý, M.; Schaminée, J.H.J.; Schwabe, A. Vegetation survey: A new focus for Applied Vegetation Science. Appl. Veg. Sci. 2011, 14, 435–439. [Google Scholar] [CrossRef]
- Landucci, F.; Acosta, A.T.R.; Agrillo, E.; Attorre, F.; Biondi, E.; Cambria, V.E.; Chiarucci, A.; Del Vico, E.; De Sanctis, M.; Facioni, L.; et al. VegItaly: The Italian collaborative project for a national vegetation database. Plant Biosyst. 2012, 146, 756–763. [Google Scholar] [CrossRef] [Green Version]
- Dengler, J.; Jansen, F.; Glöckler, F.; Peet, R.K.; De Cáceres, M.; Chytrý, M.; Ewald, J.; Oldeland, J.; Lopez-Gonzalez, G.; Finckh, M.; et al. The Global Index of Vegetation-Plot Databases (GIVD): A new resource for vegetation science. J. Veg. Sci. 2011, 22, 582–597. [Google Scholar] [CrossRef]
- Ichter, J.; Savio, L.; Evans, D.; Poncet, L. State-of-the-art of vegetation mapping in Europe: Results of a European survey and contribution to the French program CarHAB. Prodrome et Cartographie des Végétations de France 2017, 6, 335–352. [Google Scholar]
- Pesaresi, S.; Mancini, A.; Quattrini, G.; Casavecchia, S. Mapping mediterranean forest plant associations and habitats with functional principal component analysis using Landsat 8 NDVI time series. Remote Sens. 2020, 12, 1132. [Google Scholar] [CrossRef] [Green Version]
- Roelofsen, H.D.; Kooistra, L.; Van Bodegom, P.M.; Verrelst, J.; Krol, J.; Witte, J.P.M. Mapping a priori defined plant associations using remotely sensed vegetation characteristics. Remote Sens. Environ. 2014, 140, 639–651. [Google Scholar] [CrossRef]
- Corbane, C.; Lang, S.; Pipkins, K.; Alleaume, S.; Deshayes, M.; García Millán, V.E.; Strasser, T.; Vanden Borre, J.; Toon, S.; Michael, F. Remote sensing for mapping natural habitats and their conservation status–New opportunities and challenges. Int. J. Appl. Earth Obs. Geoinf. 2015, 37, 7–16. [Google Scholar] [CrossRef]
- Biondi, E.; Allegrezza, M.; Casavecchia, S.; Galdenzi, D.; Gasparri, R.; Pesaresi, S.; Soriano, P.; Tesei, G.; Blasi, C. New insight on Mediterranean and sub-Mediterranean syntaxa included in the Vegetation Prodrome of Italy. Flora Mediterr. 2015, 25, 77–102. [Google Scholar]
- Biondi, E.; Pesaresi, S.; Galdenzi, D.; Gasparri, R.; Biscotti, N.; Viscio, G.; Casavecchia, S. Post-abandonment dynamic on Mediterranean and sub-Mediterranean perennial grasslands: The edge vegetation of the new class Charybdido pancratii-Asphodeletea ramosi. Plant Sociol. 2016, 53, 3–18. [Google Scholar]
- Biondi, E.; Pesaresi, S.; Gasparri, R.; Biscotti, N.; Viscio, G.; Bonsanto, D.; Casavecchia, S. New contributions to the class Charybdido pancratii-Asphodeletea ramosi Biondi 2016. Plant Sociol. 2017, 54, 137–144. [Google Scholar]
- Schwieder, M.; Leitão, P.J.; da Cunha Bustamante, M.M.; Ferreira, L.G.; Rabe, A.; Hostert, P. Mapping Brazilian savanna vegetation gradients with Landsat time series. Int. J. Appl. Earth Obs. Geoinf. 2016, 52, 361–370. [Google Scholar] [CrossRef]
- Wessels, K.; Steenkamp, K.; von Maltitz, G.; Archibald, S. Remotely sensed vegetation phenology for describing and predicting the biomes of South Africa. Appl. Veg. Sci. 2011, 14, 49–66. [Google Scholar] [CrossRef]
- Boles, S.H.; Xiao, X.; Liu, J.; Zhang, Q.; Munkhtuya, S.; Chen, S.; Ojima, D. Land cover characterization of Temperate East Asia using multi-temporal VEGETATION sensor data. Remote Sens. Environ. 2004, 90, 477–489. [Google Scholar] [CrossRef]
- Hmimina, G.; Dufrêne, E.; Pontailler, J.-Y.; Delpierre, N.; Aubinet, M.; Caquet, B.; de Grandcourt, A.; Burban, B.; Flechard, C.; Granier, A.; et al. Evaluation of the potential of MODIS satellite data to predict vegetation phenology in different biomes: An investigation using ground-based NDVI measurements. Remote Sens. Environ. 2013, 132, 145–158. [Google Scholar] [CrossRef]
- Grabska, E.; Hostert, P.; Pflugmacher, D.; Ostapowicz, K. Forest Stand Species Mapping Using the Sentinel-2 Time Series. Remote Sens. 2019, 11, 1197. [Google Scholar] [CrossRef] [Green Version]
- Feilhauer, H.; He, K.S.; Rocchini, D. Modeling Species Distribution Using Niche-Based Proxies Derived from Composite Bioclimatic Variables and MODIS NDVI. Remote Sens. 2012, 4, 2057–2075. [Google Scholar] [CrossRef] [Green Version]
- Revermann, R.; Finckh, M.; Stellmes, M.; Strohbach, B.; Frantz, D.; Oldeland, J. Linking Land Surface Phenology and Vegetation-Plot Databases to Model Terrestrial Plant α-Diversity of the Okavango Basin. Remote Sens. 2016, 8, 370. [Google Scholar] [CrossRef] [Green Version]
- Adams, B.; Iverson, L.; Matthews, S.; Peters, M.; Prasad, A.; Hix, D.M. Mapping Forest Composition with Landsat Time Series: An Evaluation of Seasonal Composites and Harmonic Regression. Remote Sens. 2020, 12, 610. [Google Scholar] [CrossRef] [Green Version]
- Schauman, S.; Verger, A.; Filella, I.; Peñuelas, J. Characterisation of Functional-Trait Dynamics at High Spatial Resolution in a Mediterranean Forest from Sentinel-2 and Ground-Truth Data. Remote Sens. 2018, 10, 1874. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, S.; Schmitt, T.M.; Chiarucci, A.; Irl, S.D.H.; Rocchini, D.; Vetaas, O.R.; Tanase, M.A.; Mermoz, S.; Bouvet, A.; Beierkuhnlein, C. Remote sensing of β-diversity: Evidence from plant communities in a semi-natural system. Appl. Veg. Sci. 2019, 22, 13–26. [Google Scholar] [CrossRef] [Green Version]
- USA-NPN National Phenology Network Land Surface Phenology and Remote Sensing (LSP/RS). Available online: https://usanpn.org/node/14 (accessed on 13 August 2020).
- Soudani, K.; le Maire, G.; Dufrêne, E.; François, C.; Delpierre, N.; Ulrich, E.; Cecchini, S. Evaluation of the onset of green-up in temperate deciduous broadleaf forests derived from Moderate Resolution Imaging Spectroradiometer (MODIS) data. Remote Sens. Environ. 2008, 112, 2643–2655. [Google Scholar] [CrossRef]
- Soudani, K.; Hmimina, G.; Delpierre, N.; Pontailler, J.-Y.; Aubinet, M.; Bonal, D.; Caquet, B.; de Grandcourt, A.; Burban, B.; Flechard, C.; et al. Ground-based Network of NDVI measurements for tracking temporal dynamics of canopy structure and vegetation phenology in different biomes. Remote Sens. Environ. 2012, 123, 234–245. [Google Scholar] [CrossRef]
- White, M.A.; Hoffman, F.; Hargrove, W.W.; Nemani, R.R. A global framework for monitoring phenological responses to climate change. Geophys. Res. Lett. 2005, 32, L04705. [Google Scholar] [CrossRef] [Green Version]
- Bajocco, S.; Ferrara, C.; Alivernini, A.; Bascietto, M.; Ricotta, C. Remotely-sensed phenology of Italian forests: Going beyond the species. Int. J. Appl. Earth Obs. Geoinf. 2019, 74, 314–321. [Google Scholar] [CrossRef]
- Hoffman, F.M.; Kumar, J.; Hargrove, W.W. Integrating Statistical and Expert Knowledge to Develop Phenoregions for the Continental United States. In Proceedings of the AGU Fall Meeting Abstracts, San Francisco, CA, USA, 9–13 December 2013; Volume 1, p. B43C-0490. [Google Scholar]
- Morisette, J.T.; Richardson, A.D.; Knapp, A.K.; Fisher, J.I.; Graham, E.A.; Abatzoglou, J.; Wilson, B.E.; Breshears, D.D.; Henebry, G.M.; Hanes, J.M.; et al. Tracking the rhythm of the seasons in the face of global change: Phenological research in the 21st century. Front. Ecol. Environ. 2009, 7, 253–260. [Google Scholar] [CrossRef] [Green Version]
- Schaber, J.; Badeck, F.W. Physiology-based phenology models for forest tree species in Germany. Int. J. Biometeorol. 2003, 47, 193–201. [Google Scholar] [CrossRef]
- Biondi, E.; Casavecchia, S.; Gigante, D. Contribution to the syntaxonomic knowledge of the Quercus ilex L. woods of the Central European Mediterranean Basin. Fitosociologia 2003, 40, 129–156. [Google Scholar]
- Biondi, E. La Vegetazione del Monte Conero (con Carta della Vegetazione alla Scala 1:10000; Tecnostampa: Ostra Vetere, Italy, 1986. [Google Scholar]
- Poldini, L.; Sburlino, G.; Vidali, M. New syntaxonomic contribution to the Vegetation Prodrome of Italy. Plant Biosyst. 2017, 151, 1111–1119. [Google Scholar] [CrossRef]
- Pedrotti, F.; Ballelli, S.; Biondi, E.; Cortini Pedrotti, C.; Orsomando, E. Resoconto dell’escursione della Società Italiana di fitosociologia nelle Marche ed in Umbria (11–14 giugno 1979). Not. Fitosociologico 1980, 16, 73–75. [Google Scholar]
- Rivas-Martínez, S.; Sáenz, S.R.; Penas, A. Worldwide bioclimatic classification system. Glob. Geobot. 2011, 1, 1–634. [Google Scholar]
- Pesaresi, S.; Biondi, E.; Casavecchia, S. Bioclimates of Italy. J. Maps 2017, 13, 955–960. [Google Scholar] [CrossRef]
- Frontoni, E.; Mancini, A.; Zingaretti, P.; Malinverni, E.; Pesaresi, S.; Biondi, E.; Pandolfi, M.; Marseglia, M.; Sturari, M.; Zabaglia, C. SIT-REM: An Interoperable and Interactive Web Geographic Information System for Fauna, Flora and Plant Landscape Data Management. ISPRS Int. J. Geo-Inf. 2014, 3, 853–867. [Google Scholar] [CrossRef] [Green Version]
- Soenen, S.A.; Peddle, D.R.; Coburn, C.A. SCS+C: A modified Sun-canopy-sensor topographic correction in forested terrain. IEEE Trans. Geosci. Remote Sens. 2005, 43, 2148–2159. [Google Scholar] [CrossRef]
- Leutner, B.; Horning, N.; Schwalb-Willmann, J. RStoolbox: Tools for Remote Sensing Data Analysis. R Package Version 0.2.3. Available online: https://cran.r-project.org/web/packages/RStoolbox/index.html (accessed on 13 August 2020).
- Lambert, J.; Drenou, C.; Denux, J.; Balent, G.; Cheret, V. Monitoring forest decline through remote sensing time series analysis. GISci. Remote Sens. 2013, 50, 437–457. [Google Scholar] [CrossRef]
- Hyndman, R.; Athanasopoulos, G.; Bergmeir, C.; Caceres, G.; Chhay, L.; O’Hara-Wild, M.; Petropoulos, F.; Razbash, S.; Wang, E.; Yasmeen, F. Forecast: Forecasting Functions for Time Series and Linear Models. R Package Version 8.6. Available online: https://cran.r-project.org/web/packages/forecast/index.html (accessed on 13 August 2020).
- Hyndman, R.J.; Khandakar, Y. Automatic Time Series Forecasting: The forecast Package for R. J. Stat. Softw. 2008, 27, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Wood, S.N. Generalized Additive Models: An Introduction with R, 2nd ed.; Chapman and Hall/CRC: New York, NY, USA, 2017. [Google Scholar]
- Di Salvo, F.; Ruggieri, M.; Plaia, A. Functional principal component analysis for multivariate multidimensional environmental data. Environ. Ecol. Stat. 2015, 22, 739–757. [Google Scholar] [CrossRef]
- Wang, J.L.; Chiou, J.M.; Müller, H.G. Functional Data Analysis. Annu. Rev. Stat. Its Appl. 2016, 3, 257–295. [Google Scholar] [CrossRef] [Green Version]
- Jacques, J.; Preda, C. Functional data clustering: A survey. Adv. Data Anal. Classif. 2014, 8, 231–255. [Google Scholar] [CrossRef] [Green Version]
- Hurley, M.A.; Hebblewhite, M.; Gaillard, J.; Dray, S.; Taylor, K.A.; Smith, W.K.; Zager, P.; Bonenfant, C. Functional analysis of normalized difference vegetation index curves reveals overwinter mule deer survival is driven by both spring and autumn phenology. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, X.; Hadjipantelis, P.Z.; Han, K.; Ji, H. Fdapace: Functional Data Analysis and Empirical Dynamics. R Package Version 0.4.0. Available online: https://cran.r-project.org/web/packages/fdapace/index.html (accessed on 13 August 2020).
- Biondi, E.; Gubellini, L.; Pinzi, M.; Casavecchia, S. The vascular flora of Conero Regional Nature Park (Marche, Central Italy). Flora Mediterr. 2012, 22, 67–167. [Google Scholar] [CrossRef]
- De Cáceres, M.; Font, X.; Oliva, F. The management of vegetation classifications with fuzzy clustering. J. Veg. Sci. 2010, 21, 1138–1151. [Google Scholar] [CrossRef]
- Legendre, P.; Gallagher, E. Ecologically meaningful transformations for ordination of species data. Oecologia 2001, 129, 271–280. [Google Scholar] [CrossRef]
- De Cáceres, M.; Legendre, P. Associations between species and groups of sites: Indices and statistical inference. Ecology 2009, 90, 3566–3574. [Google Scholar] [CrossRef]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2.5–3. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 13 August 2020).
- Martinez Arbizu, P. pairwiseAdonis: Pairwise Multilevel Comparison Using Adonis. R Package Version 0.3. Available online: https://github.com/pmartinezarbizu/pairwiseAdonis (accessed on 3 August 2020).
- Rocchini, D. Effects of spatial and spectral resolution in estimating ecosystem α-diversity by satellite imagery. Remote Sens. Environ. 2007, 111, 423–434. [Google Scholar] [CrossRef]
- Brooks, B.-G.J.; Lee, D.C.; Pomara, L.Y.; Hargrove, W.W. Monitoring Broadscale Vegetational Diversity and Change across North American Landscapes Using Land Surface Phenology. Forests 2020, 11, 606. [Google Scholar] [CrossRef]
- Persson, M.; Lindberg, E.; Reese, H. Tree Species Classification with Multi-Temporal Sentinel-2 Data. Remote Sens. 2018, 10, 1794. [Google Scholar] [CrossRef] [Green Version]
- Bunker, B.E.; Tullis, J.A.; Cothren, J.D.; Casana, J.; Aly, M.H. Object-based Dimensionality Reduction in Land Surface Phenology Classification. AIMS Geosci. 2016, 2, 302–328. [Google Scholar] [CrossRef]
- Nguyen, T.T.H.; De Bie, C.A.J.M.; Ali, A.; Smaling, E.M.A.; Chu, T.H. Mapping the irrigated rice cropping patterns of the Mekong delta, Vietnam, through hyper-temporal SPOT NDVI image analysis. Int. J. Remote Sens. 2012, 33, 415–434. [Google Scholar] [CrossRef]
- Rocchini, D.; Butini, S.A.; Chiarucci, A. Maximizing plant species inventory efficiency by means of remotely sensed spectral distances. Glob. Ecol. Biogeogr. 2005, 14, 431–437. [Google Scholar] [CrossRef]
- Rocchini, D.; Balkenhol, N.; Carter, G.A.; Foody, G.M.; Gillespie, T.W.; He, K.S.; Kark, S.; Levin, N.; Lucas, K.; Luoto, M.; et al. Remotely sensed spectral heterogeneity as a proxy of species diversity: Recent advances and open challenges. Ecol. Inform. 2010, 5, 318–329. [Google Scholar] [CrossRef]
- Maccherini, S.; Bacaro, G.; Tordoni, E.; Bertacchi, A.; Castagnini, P.; Foggi, B.; Gennai, M.; Mugnai, M.; Sarmati, S.; Angiolini, C. Enough Is Enough? Searching for the Optimal Sample Size to Monitor European Habitats: A Case Study from Coastal Sand Dunes. Diversity 2020, 12, 138. [Google Scholar] [CrossRef] [Green Version]
- Angelini, P.; Casella, L.; Grignetti, A.; Genovesi, P. Manuali per il Monitoraggio di Specie e Habitat di Interesse Comunitario (Direttiva 92/43/CEE) in Italia: Habitat; ISPRA, Serie Manuali e Linee Guida: Rome, Italy, 2016; pp. 1–280.
- Gigante, D.; Attorre, F.; Venanzoni, R.; Acosta, A.T.R.; Agrillo, E.; Aleffi, M.; Alessi, N.; Allegrezza, M.; Angelini, P.; Angiolini, C.; et al. A methodological protocol for Annex I Habitats monitoring: The contribution of vegetation science. Plant Sociol. 2016, 53, 77–87. [Google Scholar]
- Happ, C.; Greven, S. Multivariate Functional Principal Component Analysis for Data Observed on Different (Dimensional) Domains. J. Am. Stat. Assoc. 2018, 113, 649–659. [Google Scholar] [CrossRef] [Green Version]





| Feature | Mount Conero | Mount Valmontagnana |
|---|---|---|
| Type | Coastal | Apennine |
| Area (ha) | 347 | 215 |
| Holm oak Area (ha) | 300 (86%) | 65 (30%) |
| Black hornbeam Area (ha) | 47 (14%) | 150 (70%) |
| Phytosociological relevés of holm oak | 34 (85%) | 7 (29%) |
| Phytosociological relevés of black hornbeam | 6 (15%) | 17 (71%) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pesaresi, S.; Mancini, A.; Casavecchia, S. Recognition and Characterization of Forest Plant Communities through Remote-Sensing NDVI Time Series. Diversity 2020, 12, 313. https://doi.org/10.3390/d12080313
Pesaresi S, Mancini A, Casavecchia S. Recognition and Characterization of Forest Plant Communities through Remote-Sensing NDVI Time Series. Diversity. 2020; 12(8):313. https://doi.org/10.3390/d12080313
Chicago/Turabian StylePesaresi, Simone, Adriano Mancini, and Simona Casavecchia. 2020. "Recognition and Characterization of Forest Plant Communities through Remote-Sensing NDVI Time Series" Diversity 12, no. 8: 313. https://doi.org/10.3390/d12080313
APA StylePesaresi, S., Mancini, A., & Casavecchia, S. (2020). Recognition and Characterization of Forest Plant Communities through Remote-Sensing NDVI Time Series. Diversity, 12(8), 313. https://doi.org/10.3390/d12080313