Phragmites australis Associates with Belowground Fungal Communities Characterized by High Diversity and Pathogen Abundance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Sites
2.2. Sample Collection
2.3. Molecular Methods
2.4. Statistical Analysis
3. Results
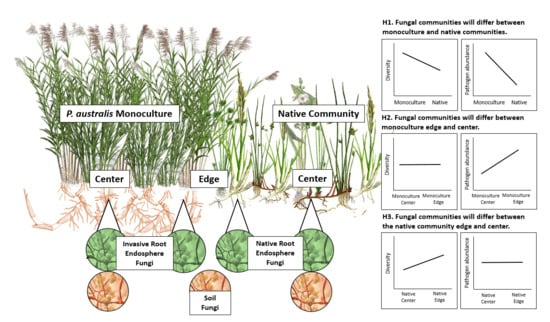
3.1. Do P. australis Monoculture Root and Soil Fungal Communities Differ from That of the Neighboring Native Community?
3.2. Does P. australis Harbor Distinct Fungal Communities at Its Expanding Edge Compared to Its Monodominant Center?
3.3. Does Proximity to the P. australis Invasion Front Alter Native Root and Soil Fungal Community Composition, Diversity or Pathogen abundances?
4. Discussion
4.1. Contrasting Fungal Composition, Richness, and Diversity in P. australis Monocultures and Native Communities
4.2. Phragmites australis Harbors Higher Putative Pathogen Abundances than Co-Occuring Native Species
4.3. Phragmites australis Harbors Distinct Soil and Endosphere Fungal Communities at Its Expanding Edge
4.4. Does Invader Presence Influence Native Endosphere Fungal Composition, Richness, Diversity, or Pathogen Abundance?
4.5. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dawson, W.; Schrama, M. Identifying the role of soil microbes in plant invasions. J. Ecol. 2016, 104, 1211–1218. [Google Scholar] [CrossRef] [Green Version]
- Van Der Putten, W.H.; Klironomos, J.N.; Wardle, D.A. Microbial ecology of biological invasions. ISME J. 2007, 1, 28–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fitzpatrick, C.R.; Copeland, J.; Wang, P.W.; Guttman, D.S.; Kotanen, P.M.; Johnson, M.T.J. Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl. Acad. Sci. USA 2018, 115, E1157–E1165. [Google Scholar] [CrossRef] [Green Version]
- Coats, V.C.; Rumpho, M.E. The rhizosphere microbiota of plant invaders: An overview of recent advances in the microbiomics of invasive plants. Front. Microbiol. 2014, 5, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Van Der Putten, W.H. Impacts of soil microbial communities on exotic plant invasions. Trends Ecol. Evol. 2010, 25, 512–519. [Google Scholar] [CrossRef]
- Reinhart, K.O.; Callaway, R.M. Soil biota and invasive plants. New Phytol. 2006, 170, 445–457. [Google Scholar] [CrossRef]
- Bever, J.D.; Westover, K.M.; Antonovics, J. Incorporating the soil community into plant population dynamics: The utility of the feedback approach. J. Ecol. 1997, 85, 561. [Google Scholar] [CrossRef]
- Aschehoug, E.T.; Callaway, R.M.; Newcombe, G.; Tharayil, N.; Chen, S. Fungal endophyte increases the allelopathic effects of an invasive forb. Oecologia 2014, 175, 285–291. [Google Scholar] [CrossRef]
- Keane, R. Exotic plant invasions and the enemy release hypothesis. Trends Ecol. Evol. 2002, 17, 164–170. [Google Scholar] [CrossRef]
- Callaway, R.M.; Cipollini, D.; Barto, K.; Thelen, G.C.; Hallett, S.G.; Prati, D.; Stinson, K.; Klironomos, J. Novel weapons: Invasive plant suppresses fungal mutualists in america but not in its native Europe. Ecology 2008, 89, 1043–1055. [Google Scholar] [CrossRef] [Green Version]
- Beckstead, J.; Parker, I.M. Invasiveness of Ammophila arenaria: Release from soil-borne pathogens? Ecology 2003, 84, 2824–2831. [Google Scholar] [CrossRef]
- Eppinga, M.B.; Rietkerk, M.; Dekker, S.C.; De Ruiter, P.C.; Van Der Putten, W.H. Accumulation of local pathogens: A new hypothesis to explain exotic plant invasions. Oikos 2006, 114, 168–176. [Google Scholar] [CrossRef] [Green Version]
- Stinson, K.A.; Campbell, S.A.; Powell, J.R.; Wolfe, B.E.; Klironomos, J.N. Invasive plant suppresses the growth of native tree seedlings by disrupting belowground mutualisms. PLoS Biol. 2006, 4, e140. [Google Scholar] [CrossRef] [PubMed]
- Suding, K.N.; Harpole, W.S.; Fukami, T.; Kulmatiski, A.; MacDougall, A.S.; Stein, C.; Van Der Putten, W.H. Consequences of plant-soil feedbacks in invasion. J. Ecol. 2013, 101, 298–308. [Google Scholar] [CrossRef]
- Klironomos, J.N. Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature 2002, 417, 67–70. [Google Scholar] [CrossRef]
- Rodríguez-Caballero, G.; Roldán, A.; Caravaca, F. Invasive Nicotiana glauca shifts the soil microbial community composition and functioning of harsh and disturbed semiarid Mediterranean environments. Biol. Invasions 2020, 22, 2923–2940. [Google Scholar] [CrossRef]
- Gomes, S.I.F.; Merckx, V.S.F.T.; Hynson, N.A. Biological invasions increase the richness of arbuscular mycorrhizal fungi from a Hawaiian subtropical ecosystem. Biol. Invasions 2018, 20, 2421–2437. [Google Scholar] [CrossRef] [Green Version]
- Clay, K.; Shearin, Z.R.C.; Bourke, K.A.; Bickford, W.A.; Kowalski, K.P. Diversity of fungal endophytes in non-native Phragmites australis in the Great Lakes. Biol. Invasions 2016, 18, 2703–2716. [Google Scholar] [CrossRef]
- Soares, M.A.; Li, H.; Kowalski, K.P. Evaluation of the functional roles of fungal endophytes of Phragmites australis from high saline and low saline habitats. Biol. Invasions 2016, 18, 2689–2702. [Google Scholar] [CrossRef]
- Shipunov, A.; Newcombe, G.; Raghavendra, A.K.H.; Anderson, C.L. Hidden diversity of endophytic fungi in an invasive plant. Am. J. Bot. 2008, 95, 1096–1108. [Google Scholar] [CrossRef]
- Allen, W.J.; Devries, A.E.; Bologna, N.J.; Bickford, W.A.; Kowalski, K.P.; Meyerson, L.A.; Cronin, J.T. Intraspecific and biogeographical variation in foliar fungal communities and pathogen damage of native and invasive Phragmites australis. Glob. Ecol. Biogeogr. 2020, 29, 1199–1211. [Google Scholar] [CrossRef]
- Bickford, W.A.; Goldberg, D.E.; Kowalski, K.P.; Zak, D.R. Root endophytes and invasiveness: No difference between native and non-native Phragmites in the Great Lakes Region. Ecosphere 2018, 9, e02526. [Google Scholar] [CrossRef] [Green Version]
- Bowen, J.L.; Kearns, P.J.; Byrnes, J.E.K.; Wigginton, S.; Allen, W.J.; Greenwood, M.; Tran, K.; Yu, J.; Cronin, J.T.; Meyerson, L.A. Lineage overwhelms environmental conditions in determining rhizosphere bacterial community structure in a cosmopolitan invasive plant. Nat. Commun. 2017, 8, 433. [Google Scholar] [CrossRef] [PubMed]
- Nelson, E.B.; Karp, M.A. Soil pathogen communities associated with native and non-native Phragmites australis populations in freshwater wetlands. Ecol. Evol. 2013, 3, 5254–5267. [Google Scholar] [CrossRef] [PubMed]
- Dean, S.L.; Farrer, E.C.; Taylor, D.L.; Porras-Alfaro, A.; Suding, K.N.; Sinsabaugh, R.L. Nitrogen deposition alters plant-fungal relationships: Linking belowground dynamics to aboveground vegetation change. Mol. Ecol. 2013, 23, 1364–1378. [Google Scholar] [CrossRef] [PubMed]
- Cesarino, I.; Ioio, R.D.; Kirschner, G.K.; Ogden, M.S.; Picard, K.L.; Rast-Somssich, M.I.; Somssich, M. Plant science’s next top models. Ann. Bot. 2020, 126, 1–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clevering, O.A.; Lissner, J. Taxonomy, chromosome numbers, clonal diversity and population dynamics of Phragmites australis. Aquat. Bot. 1999, 64, 185–208. [Google Scholar] [CrossRef]
- Meyerson, L.A.; Cronin, J.T.; Pyšek, P. Phragmites australis as a model organism for studying plant invasions. Biol. Invasions 2016, 18, 2421–2431. [Google Scholar] [CrossRef]
- Achenbach, L.; Brix, H. Can differences in salinity tolerance explain the distribution of four genetically distinct lineages of Phragmites australis in the Mississippi River Delta? Hydrobiologia 2013, 737, 5–23. [Google Scholar] [CrossRef]
- Mozdzer, T.J.; Brisson, J.; Hazelton, E.L.G. Physiological ecology and functional traits of North American native and Eurasian introduced Phragmites australis lineages. AoB Plants 2013, 5, plt048. [Google Scholar] [CrossRef]
- Vasquez, E.; Glenn, E.; Brown, J.; Guntenspergen, G.; Nelson, S. Salt tolerance underlies the cryptic invasion of North American salt marshes by an introduced haplotype of the common reed Phragmites australis (Poaceae). Mar. Ecol. Prog. Ser. 2005, 298, 1–8. [Google Scholar] [CrossRef]
- Fischer, M.S.; Rodriguez, R.J. Fungal endophytes of invasive Phagramites australis populations vary in species composition and fungicide susceptibility. Symbiosis 2013, 61, 55–62. [Google Scholar] [CrossRef]
- Saltonstall, K. Cryptic invasion by a non-native genotype of the common reed, Phragmites australis, into North America. Proc. Natl. Acad. Sci. USA 2002, 99, 2445–2449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambertini, C.; Mendelssohn, I.A.; Gustafsson, M.H.G.; Olesen, B.; Riis, T.; Sorrell, B.K.; Brix, H. Tracing the origin of Gulf Coast Phragmites (Poaceae): A story of long-distance dispersal and hybridization. Am. J. Bot. 2012, 99, 538–551. [Google Scholar] [CrossRef]
- Meyerson, L.A.; Lambertini, C.; McCormick, M.K.; Whigham, D.F. Hybridization of common reed in North America? The answer is blowing in the wind. AoB Plants 2012, 2012, pls 002. [Google Scholar] [CrossRef] [PubMed]
- Meyerson, L.A.; Lambert, A.M.; Saltonstall, K. A Tale of three lineages: Expansion of common reed (Phragmites australis) in the U.S. Southwest and Gulf Coast. Invasive Plant. Sci. Manag. 2010, 3, 515–520. [Google Scholar] [CrossRef]
- Farrer, E.C.; Birnbaum, C.; Waryszak, P.; Halbrook, S.R.; Brady, M.V.; Bumby, C.R.; Candaele, H.; Kulick, D.; Lee, S.F.H.; Schroeder, C.S.; et al. Above and belowground impacts of an invasive species vary across the landscape. J. Ecol. 2016. under review. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Hill, M.O. Diversity and Evenness: A Unifying Notation and Its Consequences. Ecology 1973, 54, 427–432. [Google Scholar] [CrossRef] [Green Version]
- Alberdi, A.; Gilbert, M.T.P. Hilldiv: An r package for the integral analysis of diversity based on Hill numbers. bioRxiv 2019, 545665. [Google Scholar] [CrossRef] [Green Version]
- Jost, L. Partitioning diversity into independent alpha and beta components. Ecology 2007, 88, 2427–2439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.G.; Simpson, L.; Solymos, P.; et al. Vegan: Community Ecology Package; R package Version 2.5-5; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D.; R Core Team. Nlme: Linear and Non-Linear Mixed Effects Models; EcoLab University of Granada: Granada, Spain, 2019. [Google Scholar]
- Zuur, A.F.; Ieno, E.N.; Walker, N.; Saveliev, A.A.; Smith, G.M. Mixed Effects Models and Extensions in Ecology with R.; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Borcard, D.; Legendre, P. All-scale spatial analysis of ecological data by means of principal coordinates of neighbour matrices. Ecol. Model. 2002, 153, 51–68. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- McMurdie, P.J.; Holmes, S. phyloseq: An r package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenth, R. emmeans: Estimated Marginal Means, aka Least-Squares Means; University of Iowa: Iowa City, IA, USA, 2020. [Google Scholar]
- Kirk, P.M.; Cannon, P.F.; Minter, D.W.; Stalpers, J.A. Dictionary of the Fungi, 10th ed.; CAB International: Wallingford, UK, 2008. [Google Scholar]
- Westover, K.M.; Kennedy, A.C.; Kelley, S.E. Patterns of rhizosphere microbial community structure associated with co-occurring plant species. J. Ecol. 1997, 85, 863. [Google Scholar] [CrossRef]
- Rosenblueth, M.; Martinez-Romero, E. Bacterial endophytes and their interactions with hosts. Mol. Plant. Microbe Interact. 2006, 19, 827–837. [Google Scholar] [CrossRef] [Green Version]
- Hardoim, P.R.; Van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Boil. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [Green Version]
- Micallef, S.A.; Shiaris, M.P.; Colón-Carmona, A. Influence of Arabidopsis thaliana accessions on rhizobacterial communities and natural variation in root exudates. J. Exp. Bot. 2009, 60, 1729–1742. [Google Scholar] [CrossRef] [Green Version]
- Olivares, F.L.; Baldani, V.L.D.; Reis, V.M.; Baldani, J.I.; Döbereiner, J. Occurrence of the endophytic diazotrophs Herbaspirillum spp. in roots, stems and leaves predominantly of Gramineae. Biol. Fertil. Soils 1996, 21, 197–200. [Google Scholar] [CrossRef]
- Weber, O.; Baldani, V.; Teixeira, K.; Kirchhof, G.; Baldani, J.; Döbereiner, J. Isolation and characterization of diazotrophic bacteria from banana and pineapple plants. Plant. Soil 1999, 210, 103–113. [Google Scholar] [CrossRef]
- Si, C.; Liu, X.; Wang, C.; Wang, L.; Dai, Z.; Qi, S.; Du, D. Different degrees of plant invasion significantly affect the richness of the soil fungal community. PLoS ONE 2013, 8, e85490. [Google Scholar] [CrossRef] [PubMed]
- Lankau, R. Resistance and recovery of soil microbial communities in the face of Allaria petiolata invasions. New Phytol. 2011, 189, 536–548. [Google Scholar] [CrossRef] [PubMed]
- Mangla, S.; Callaway, R.M. Exotic invasive plant accumulates native soil pathogens which inhibit native plants. J. Ecol. 2007, 96, 58–67. [Google Scholar] [CrossRef]
- Nijjer, S.; Rogers, W.E.; Siemann, E. Negative plant–soil feedbacks may limit persistence of an invasive tree due to rapid accumulation of soil pathogens. Proc. R Soc. B Boil. Sci. 2007, 274, 2621–2627. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Zhou, J.; Zeng, T.; Miao, Y.; Mei, L.; Yao, G.; Fang, K.; Dong, X.; Sha, T.; Yang, M.; et al. Quantifying the sharing of foliar fungal pathogens by the invasive plant Ageratina adenophora and its neighbours. New Phytol. 2020, 227, 1493–1504. [Google Scholar] [CrossRef]
- Diez, J.M.; Dickie, I.; Edwards, G.R.; Hulme, P.E.; Sullivan, J.J.; Duncan, R.P. Negative soil feedbacks accumulate over time for non-native plant species. Ecol. Lett. 2010, 13, 803–809. [Google Scholar] [CrossRef]
- Flory, S.L.; Clay, K. Pathogen accumulation and long-term dynamics of plant invasions. J. Ecol. 2013, 101, 607–613. [Google Scholar] [CrossRef]
- Elton, C.S. The Ecology of Invasions by Animals and Plants; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 1958. [Google Scholar]
- Levine, J.; Pachepsky, E.; Kendall, B.E.; Yelenik, S.; Lambers, J.H.R. Plant-soil feedbacks and invasive spread. Ecol. Lett. 2006, 9, 1005–1014. [Google Scholar] [CrossRef]
- Devries, A.E.; Kowalski, K.P.; Bickford, W.A. Growth and behavior of North American microbes on Phragmites australis leaves. Microorganisms 2020, 8, 690. [Google Scholar] [CrossRef]
- Allen, W.J.; Meyerson, L.A.; Flick, A.J.; Cronin, J.T. Intraspecific variation in indirect plant-soil feedbacks influences a wetland plant invasion. Ecology 2018, 99, 1430–1440. [Google Scholar] [CrossRef]
- Castrillo, G.; Teixeira, P.J.P.L.; Paredes, S.H.; Law, T.F.; De Lorenzo, L.; Feltcher, M.E.; Finkel, O.M.; Breakfield, N.W.; Mieczkowski, P.A.; Jones, C.D.; et al. Root microbiota drive direct integration of phosphate stress and immunity. Nature 2017, 543, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Hacquard, S.; Garrido-Oter, R.; González, A.; Spaepen, S.; Ackermann, G.; Lebeis, S.; McHardy, A.C.; Dangl, J.L.; Knight, R.; Ley, R.; et al. Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe 2015, 17, 603–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Power, A.G.; Mitchell, C.E. Pathogen spillover in disease epidemics. Am. Nat. 2004, 164, S79–S89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonsall, M.B.; Hassell, M.P. Apparent competition structures ecological assemblages. Nature 1997, 388, 371–373. [Google Scholar] [CrossRef]
- Lekberg, Y.; Vasar, M.; Bullington, L.S.; Sepp, S.; Antunes, P.M.; Bunn, R.A.; Larkin, B.G.; Öpik, M. More bang for the buck? Can arbuscular mycorrhizal fungal communities be characterized adequately alongside other fungi using general fungal primers? New Phytol. 2018, 220, 971–976. [Google Scholar] [CrossRef] [Green Version]
- Redman, R.S.; Dunigan, D.D.; Rodriguez, R. Fungal symbiosis from mutualism to parasitism: Who controls the outcome, host or invader? New Phytol. 2001, 151, 705–716. [Google Scholar] [CrossRef] [Green Version]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schroeder, C.S.; Halbrook, S.; Birnbaum, C.; Waryszak, P.; Wilber, W.; Farrer, E.C. Phragmites australis Associates with Belowground Fungal Communities Characterized by High Diversity and Pathogen Abundance. Diversity 2020, 12, 363. https://doi.org/10.3390/d12090363
Schroeder CS, Halbrook S, Birnbaum C, Waryszak P, Wilber W, Farrer EC. Phragmites australis Associates with Belowground Fungal Communities Characterized by High Diversity and Pathogen Abundance. Diversity. 2020; 12(9):363. https://doi.org/10.3390/d12090363
Chicago/Turabian StyleSchroeder, Carolyn S., Susannah Halbrook, Christina Birnbaum, Paweł Waryszak, William Wilber, and Emily C. Farrer. 2020. "Phragmites australis Associates with Belowground Fungal Communities Characterized by High Diversity and Pathogen Abundance" Diversity 12, no. 9: 363. https://doi.org/10.3390/d12090363
APA StyleSchroeder, C. S., Halbrook, S., Birnbaum, C., Waryszak, P., Wilber, W., & Farrer, E. C. (2020). Phragmites australis Associates with Belowground Fungal Communities Characterized by High Diversity and Pathogen Abundance. Diversity, 12(9), 363. https://doi.org/10.3390/d12090363