Diversity of Endophytic and Pathogenic Fungi of Saffron (Crocus sativus) Plants from Cultivation Sites in Italy
Abstract
:1. Introduction
2. Materials and Methods
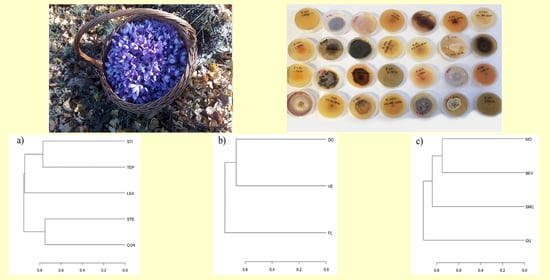
2.1. Biological Material and Study Sites
2.2. Isolation of Fungi and Molecular Identification
2.3. Phylogenetic Analysis
2.4. Diversity Analyses
3. Results
3.1. Isolation and Identification of Saffron Endophytic Fungi
3.2. Endophytic Fungi from Healthy Plants
3.3. Isolated Fungi from Rotten Plants
4. Discussion
4.1. Biodiversity of Saffron Fungal Endophytes
4.2. Potential Biotechnological Importance of Saffron Endophytes
4.3. Plant Growth-Promoting, Pathogenic, and Anti-Pathogenic Effects of Saffron-Associated Fungi
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carroll, G.C. The biology of endophytism in plants with particular reference to woody plants. In Microbiology of the Phyllosphere; Fokkema, N.J., van den Heuvel, J., Eds.; Cambridge University Press: Cambridge, UK, 1986; pp. 205–222. [Google Scholar]
- Petrini, O. Fungal Endophytes of Tree Leaves. In Microbial Ecology of Leaves; Andrews, J.H., Hirano, S.S., Eds.; Springer: New York, NY, USA, 1991; pp. 179–197. [Google Scholar]
- Arnold, A.E. Understanding the Diversity of Foliar Endophytic Fungi: Progress, Challenges, and Frontiers. Fungal Biol. Rev. 2007, 21, 51–66. [Google Scholar] [CrossRef]
- Stierle, A.; Strobel, G.; Stierle, D.; Grothaus, P.; Bignami, G. The Search for a Taxol-Producing Microorganism among the Endophytic Fungi of the Pacific Yew, Taxus Brevifolia. J. Nat. Prod. 1995, 58, 1315–1324. [Google Scholar] [CrossRef] [PubMed]
- Nisa, H.; Kamili, A.N.; Nawchoo, I.A.; Shafi, S.; Shameem, N.; Bandh, S.A. Fungal Endophytes as Prolific Source of Phytochemicals and Other Bioactive Natural Products: A Review. Microb. Pathog. 2015, 82, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, K.R. Diversity, Ecology, and Significance of Fungal Endophytes. In Endophytes and Secondary Metabolites; Jha, S., Ed.; Springer: Cham, Switzerland, 2019; pp. 61–100. [Google Scholar] [CrossRef]
- Zheng, Y.K.; Qiao, X.-G.; Miao, C.P.; Liu, K.; Chen, Y.W.; Xu, L.H.; Zhao, L.X. Diversity, Distribution and Biotechnological Potential of Endophytic Fungi. Ann. Microbiol. 2016, 66, 529–542. [Google Scholar] [CrossRef]
- De Silva, N.I.; Brooks, I.; Lumyong, S.; Hyde, K.D. Use of endophytes as biocontrol agents. Fungal Biol. Rev. 2019, 33, 133–148. [Google Scholar] [CrossRef]
- Mathew, B. The Crocuses: A revision of the Genus Crocus (Iridaceous); Batsford, B.T.: London, UK, 1982. [Google Scholar]
- Cardone, L.; Castronuovo, D.; Perniola, M.; Cicco, N.; Candido, V. Saffron (Crocus sativus L.), the king of spices: An overview. Sci. Hortic. 2020, 272, 109560. [Google Scholar] [CrossRef]
- Schmidt, T.; Heitkam, T.; Liedtke, S.; Schubert, V.; Menzel, G. Adding Color to a Century-Old Enigma: Multi-Color Chromosome Identification Unravels the Autotriploid Nature of Saffron (Crocus sativus) as a Hybrid of Wild Crocus Cartwrightianus Cytotypes. New Phytol. 2019, 222, 1965–1980. [Google Scholar] [CrossRef]
- Nemati, Z.; Harpke, D.; Gemicioglu, A.; Kerndorff, H.; Blattner, F.R. Saffron (Crocus sativus) Is an Autotriploid That Evolved in Attica (Greece) from Wild Crocus Cartwrightianus. Mol. Phylogenet. Evol. 2019, 136, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Basker, D.; Negbi, M. Uses of Saffron. Econ. Bot. 1983, 37, 228–236. [Google Scholar] [CrossRef]
- Gresta, F.; Lombardo, G.M.; Siracusa, L.; Ruberto, G. Saffron, an Alternative Crop for Sustainable Agricultural Systems. A Review. Agron. Sustain. Dev. 2008, 28, 95–112. [Google Scholar] [CrossRef] [Green Version]
- Shawky, E.; Abu El-Khair, R.M.; Selim, D.A. NIR Spectroscopy-Multivariate Analysis for Rapid Authentication, Detection and Quantification of Common Plant Adulterants in Saffron (Crocus sativus L.) Stigmas. LWT 2020, 122, 109032. [Google Scholar] [CrossRef]
- Deka, D.; Tayung, K.; Jha, D.K. Harnessing Fungal Endophytes for Plant and Human Health. In Endophytes: Biology and Biotechnology; Maheshwari, D.K., Ed.; Springer: Cham, Switzerland, 2017; Volume 1, pp. 59–98. [Google Scholar] [CrossRef]
- Raj, P.; Khan, S.; Modak, M.; Lone, Z.; Rather, S.; Yaqoob, M. Biodiversity of Endophytic Fungi in Saffron (Crocus sativus) Andantimicrobial Activity of Their Crude Extract. Indo Am. J. Pharm. 2013, 3, 3702–3713. [Google Scholar]
- Wani, Z.A.; Mirza, D.N.; Arora, P.; Riyaz-Ul-Hassan, S. Molecular Phylogeny, Diversity, Community Structure, and Plant Growth Promoting Properties of Fungal Endophytes Associated with the Corms of Saffron Plant: An Insight into the Microbiome of Crocus Sativus Linn. Fungal Biol. 2016, 120, 1509–1524. [Google Scholar] [CrossRef] [PubMed]
- Ambardar, S.; Singh, H.R.; Gowda, M.; Vakhlu, J. Comparative Metagenomics Reveal Phylum Level Temporal and Spatial Changes in Mycobiome of Belowground Parts of Crocus sativus. PLoS ONE 2016, 11, e0163300. [Google Scholar] [CrossRef] [Green Version]
- Chamkhi, I.; Sbabou, L.; Aurag, J. Endophytic Fungi Isolated from Crocus Sativus, L. (Saffron) as a Source of Bioactive Secondary Metabolites. Pharmacogn. J. 2018, 10, 1143–1148. [Google Scholar] [CrossRef] [Green Version]
- Jan, B.; Reshi, Z.A.; Mohiddin, F.A. Correction to: Site and Organ-Specific Culture-Dependent Endophytic Diversity of Crocus sativus L. (Saffron) in Kashmir Himalaya, India. Microb. Ecol. 2021. [Google Scholar] [CrossRef]
- Victorino, Í.M.M.; Voyron, S.; Caser, M.; Orgiazzi, A.; Demasi, S.; Berruti, A.; Scariot, V.; Bianciotto, V.; Lumini, E. Metabarcoding of Soil Fungal Communities Associated with Alpine Field-Grown Saffron (Crocus sativus L.) Inoculated with AM Fungi. J. Fungi 2021, 7, 45. [Google Scholar] [CrossRef]
- Rodrigues, K.F. The Foliar Fungal Endophytes of the Amazonian Palm Euterpe Oleracea. Mycologia 1994, 86, 376–385. [Google Scholar] [CrossRef]
- Sanei, S.J.; Razavi, S.E. In vitro antifungal activities of saffron (Crocus sativus L.) stigmas against Aspergillus species and toxin production. Iran. J. Med. Aromat. Plants Res. 2018, 34, 77–86. [Google Scholar]
- Khoulati, A.; Ouahhoud, S.; Mamri, S.; Alaoui, K.; Lahmass, I.; Choukri, M.; Kharmach, E.Z.; Asehraou, A.; Saalaoui, E. Saffron extract stimulates growth, improves the antioxidant components of Solanum lycopersicum L., and has an antifungal effect. Ann. Agric. Sci. 2019, 64, 138–150. [Google Scholar] [CrossRef]
- Arnold, A.E.; Lutzoni, F. Diversity and Host Range of Foliar Fungal Endophytes: Are Tropical Leaves Biodiversity Hotspots? Ecology 2007, 88, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylo-genetics. In PCR Protocols. A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Rubini, A.; Paolocci, F.; Granetti, B.; Arcioni, S. Single Step Molecular Characterization of Morphologically Similar Black Truffle Species. FEMS Microbiol. Lett. 1998, 164, 7–12. [Google Scholar] [CrossRef]
- Hall, A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Huang, Y.; Niu, B.; Gao, Y.; Fu, L.; Li, W. CD-HIT Suite: A Web Server for Clustering and Comparing Biological Sequences. Bioinformatics 2010, 26, 680–682. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Rivera-Orduña, F.N.; Suarez-Sanchez, R.A.; Flores-Bustamante, Z.R.; Gracida-Rodriguez, J.N.; Flores-Cotera, L.B. Diversity of Endophytic Fungi of Taxus Globosa (Mexican Yew). Fungal Divers. 2011, 47, 65–74. [Google Scholar] [CrossRef]
- Jaccard, P. Lois de Distribution Florale Dans La Zone Alpine. Bull. Soc. Vaudoise Sci. Nat. 1902, 38, 69–130. [Google Scholar]
- Sorensen, T.A. A method of establishing groups of equal amplitude in plant sociology based on similarity of species content and its application to analyses of the vegetation on Danish commons. Biol. Skar. 1948, 5, 1–34. [Google Scholar]
- Anderson, M.J. A New Method for Non-Parametric Multivariate Analysis of Variance. Austral. Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An Open Annotation Tool for Parsing Fungal Community Datasets by Ecological Guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Cappelli, C. Occurrence of Fusarium oxysporum f. sp. gladioli on saffron in Italy. Phytopathol. Mediterr. 1994, 33, 93–94. [Google Scholar]
- Ghosh, B.; Ray, R.R. Current commercial perspective of Rhizopus oryzae: A review. J. Appl. Sci. 2011, 11, 2470–2486. [Google Scholar] [CrossRef] [Green Version]
- Medina-Córdova, N.; López-Aguilar, R.; Ascencio, F.; Castellanos, T.; Campa-Córdova, A.I.; Angulo, C. Biocontrol Activity of the Marine Yeast Debaryomyces Hansenii against Phytopathogenic Fungi and Its Ability to Inhibit Mycotoxins Production in Maize Grain (Zea mays L.). Biol. Control 2016, 97, 70–79. [Google Scholar] [CrossRef]
- Martini, M.; Musetti, R.; Grisan, S.; Polizzotto, R.; Borselli, S.; Pavan, F.; Osler, R. DNA-Dependent Detection of the Grapevine Fungal Endophytes Aureobasidium Pullulans and Epicoccum Nigrum. Plant Dis. 2009, 93, 993–998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bae, H.; Roberts, D.P.; Lim, H.S.; Strem, M.D.; Park, S.C.; Ryu, C.M.; Melnick, R.L.; Bailey, B.A. Endophytic Trichoderma Isolates from Tropical Environments Delay Disease Onset and Induce Resistance Against Phytophthora Capsici in Hot Pepper Using Multiple Mechanisms. Mol. Plant Microbe Interact. 2011, 24, 336–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazerooni, E.A.; Rethinasamy, V.; Al-Sadi, A.M. Talaromyces Pinophilus Inhibits Pythium and Rhizoctonia-Induced Damping-off of Cucumber. J. Plant Pathol. 2019, 101, 377–383. [Google Scholar] [CrossRef]
- Granito, V.M.; Lunghini, D.; Maggi, O.; Persiani, A.M. Wood- inhabiting fungi in southern Italy forest stands: Morphogroups, vegeta-tion types and decay classes. Mycologia 2015, 107, 1074–1088. [Google Scholar] [CrossRef] [PubMed]
- Li, W.C.; Zhou, J.; Guo, S.Y.; Guo, L.D. Endophytic fungi associated with lichens in Baihua mountain of Beijing, China. Fungal Divers. 2007, 25, 69–80. [Google Scholar]
- Costa, I.P.M.W.; Assuncao, M.M.C.; Lima, T.E.F.; Oliveira, R.J.V.; Cavalcanti, M.A.Q. Checklist of endophytic fungi from tropical regions. Mycotaxon 2012, 119, 494. [Google Scholar]
- Widmer, T.L.; McMahon, M.B.; Luster, D.G. Plant pathogenic fungi are harbored as endophytes in Rhododendron spp. native to the Eastern USA. Fungal Ecol. 2020, 47, 100949. [Google Scholar] [CrossRef]
- Lambert, C.; Pourmoghaddam, M.J.; Cedeño-Sanchez, M.; Surup, F.; Khodaparast, S.A.; Krisai-Greilhuber, I.; Voglmayr, H.; Stradal, T.E.B.; Stadler, M. Resolution of the Hypoxylon fuscum Complex (Hypoxylaceae, Xylariales) and Discovery and Biological Characterization of Two of Its Prominent Secondary Metabolites. J. Fungi 2021, 7, 131. [Google Scholar] [CrossRef]
- De Errasti, A.; Carmarán, C.C.; Novas, M.V. Diversity and significance of fungal endophytes from living stems of naturalized trees from Argentina. Fungal Divers. 2010, 411, 29–40. [Google Scholar] [CrossRef]
- De Silva, N.I.; Maharachchikumbura, S.S.N.; Thambugala, K.M.; Bhat, D.J.; Karunarathna, S.C.; Tennakoon, D.S.; Phookamsak, R.; Jayawardena, R.S.; Lumyong, S.; Hyde, K.D. Morpho-molecular taxonomic studies reveal a high number of endophytic fungi from Magnolia candolli and M. garrettii in China and Thailand. Mycosophere 2021, 12, 163–237. [Google Scholar]
- Khusnullina, A.I.; Bilanenko, E.N.; Kurakov, A.V. Microscopic Fungi of White Sea Sediments. Contemp. Prob. Ecol. 2018, 11, 503–513. [Google Scholar] [CrossRef]
- Ponizovskaya, V.B.; Rebrikova, N.L.; Kachalkin, A.V.; Antropova, A.B.; Bilanenko, E.N.; Mokeeva, V.L. Micromycetes as Colonizers of Mineral Building Materials in Historic Monuments and Museums. Fungal Biol. 2019, 123, 290–306. [Google Scholar] [CrossRef] [PubMed]
- Tsang, C.C.; Chan, J.F.W.; Pong, W.M.; Chen, J.H.K.; Ngan, A.H.Y.; Cheung, M.; Lai, C.K.C.; Tsang, D.N.C.; Lau, S.K.P.; Woo, P.C.Y. Cutaneous Hyalohyphomycosis Due to Parengyodontium Album Gen. et Comb. Nov. Med. Mycol. J. 2016, 54, 699–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.; Yang, H.Y.; You, X.L.; Li, Y.H. Diversity of endophytic fungi from roots of Panax ginseng and their saponin yield capacities. SpringerPlus 2013, 2, 1–9. [Google Scholar]
- Seifert, K.; Hoekstra, E.; Frisvad, J.; Louis-Seize, G. Penicillium Cecidicola, a New Species on Cynipid Insect Galls on Quercus Pacifica in the Western United States. Stud. Mycol. 2004, 50, 517–523. [Google Scholar]
- Hubka, V.; Nováková, A.; Samson, R.A.; Houbraken, J.; Frisvad, J.C.; Sklenář, F.; Varga, J.; Kolařík, M. Aspergillus Europaeus Sp. Nov., a Widely Distributed Soil-Borne Species Related to A. Wentii (Section Cremei). Plant Syst. Evol. 2016, 302, 641–650. [Google Scholar] [CrossRef]
- Day, M.J.; Currah, R.S. Role of Selected Dark Septate Endophyte Species and Other Hyphomycetes as Saprobes on Moss Gametophytes. Botany 2011, 89, 349–359. [Google Scholar] [CrossRef]
- Likar, M.; Regvar, M. Isolates of Dark Septate Endophytes Reduce Metal Uptake and Improve Physiology of Salix caprea L. Plant Soil 2013, 370, 593–604. [Google Scholar] [CrossRef]
- Grünig, C.R.; Queloz, V.; Sieber, T.N. Structure of Diversity in Dark Septate Endophytes: From Species to Genes. In Endophytes of Forest Trees: Biology and Applications; Pirttilä, A.M., Frank, A.C., Eds.; Springer: Dordrecht, NL, USA, 2011; pp. 3–30. [Google Scholar] [CrossRef]
- Jumpponen, A.; Trappe, J.M. Dark Septate Endophytes: A Review of Facultative Biotrophic Root-Colonizing Fungi. New Phytol. 1998, 140, 295–310. [Google Scholar] [CrossRef]
- Sieber, T.N.; Grünig, C.R. Fungal root endophytes. In Plant Roots—The Hidden Half; Eshel, A., Beeckman, T., Eds.; CRC Press: Boca Raton, FL, USA, 2013; pp. 1–38. [Google Scholar]
- Butler, M.J.; Day, A.W. Fungal Melanins: A Review. Can. J. Microbiol. 1998, 44, 1115–1136. [Google Scholar] [CrossRef]
- Eisenman, H.C.; Casadevall, A. Synthesis and Assembly of Fungal Melanin. Appl. Microbiol. Biotechnol. 2012, 93, 931–940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berthelot, C.; Perrin, Y.; Leyval, C.; Blaudez, D. Melanization and Ageing Are Not Drawbacks for Successful Agro-Transformation of Dark Septate Endophytes. Fungal Biol. 2017, 121, 652–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandyam, K.; Jumpponen, A. Seeking the Elusive Function of the Root-Colonising Dark Septate Endophytic Fungi. Stud. Mycol. 2005, 53, 173–189. [Google Scholar] [CrossRef] [Green Version]
- Newsham, K.K. A Meta-Analysis of Plant Responses to Dark Septate Root Endophytes. New Phytol. 2011, 190, 783–793. [Google Scholar] [CrossRef]
- Kumar, D.S.S.; Hyde, K.D. Biodiversity and tissue-recurrence of endophytic fungi in Tripterygium wilfordii. Fungal Divers. 2004, 17, 69–90. [Google Scholar]
- Bukhari, S.; Din, I.; Grewal, S.; Dhar, M. Antiproliferative Effect of Saffron and Its Constituents on Different Cancerous Cell Lines. Pharmacogn. Res. 2018, 10, 291–295. [Google Scholar] [CrossRef]
- Wani, Z.A.; Kumar, A.; Sultan, P.; Bindu, K.; Riyaz-Ul-Hassan, S.; Ashraf, N. Mortierella alpina CS10E4, an oleaginous fungal endophyte of Crocus sativus L. enhances apocarotenoid biosynthesis and stress tolerance in the host plant. Sci. Rep. 2017, 7, 8598. [Google Scholar] [CrossRef] [PubMed]
- Li, C.X.; Zhao, S.; Zhang, T.; Xian, L.; Liao, L.S.; Liu, J.L.; Feng, J.X. Genome Sequencing and Analysis of Talaromyces Pinophilus Provide Insights into Biotechnological Applications. Sci. Rep. 2017, 7, 490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinale, F.; Nicoletti, R.; Lacatena, F.; Marra, R.; Sacco, A.; Lombardi, N.; d’Errico, G.; Digilio, M.C.; Lorito, M.; Woo, S.L. Secondary Metabolites from the Endophytic Fungus Talaromyces Pinophilus. Nat. Prod. Res. 2017, 31, 1778–1785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aït Hamza, M.; Lakhtar, H.; Tazi, H.; Moukhli, A.; Fossati-Gaschignard, O.; Miché, L.; Roussos, S.; Ferji, Z.; El Mousadik, A.; Mateille, T.; et al. Diversity of Nematophagous Fungi in Moroccan Olive Nurseries: Highlighting Prey-Predator Interactions and Efficient Strains against Root-Knot Nematodes. Biol. Control 2017, 114, 14–23. [Google Scholar] [CrossRef]
- Deka, D.; Jha, D.K. Bioactivity Assessment of Endophytic Fungi Associated with Citrus Macroptera Montr.: An Endangered Ethnomedicinal Plant Used in Folk Medicines in North-East India. Indian Phytopathol. 2020, 73, 21–33. [Google Scholar] [CrossRef]
- Ahrazem, O.; Rubio-Moraga, A.; Castillo-López, R.; Trapero, A.; Gómez-Gómez, L. Crocus sativus pathogens and defence responses. In Functional Plant Science and Biotechnology, Special Issue Saffron; Husaini, A.M., Ed.; Global Science Books: London, UK, 2010; pp. 81–90. [Google Scholar]
- Bogner, C.W.; Kamdem, R.S.T.; Sichtermann, G.; Matthäus, C.; Hölscher, D.; Popp, J.; Proksch, P.; Grundler, F.M.W.; Schouten, A. Bioactive Secondary Metabolites with Multiple Activities from a Fungal Endophyte. Microb. Biotechnol. 2017, 10, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Fatima, N.; Ismail, T.; Muhammad, S.A.; Jadoon, M.; Ahmed, S.; Azhar, S.; Mumtaz, A. Epicoccum sp., an emerging source of unique bioactive metabolites. Acta Pol. Pharm. 2016, 73, 13–21. [Google Scholar] [PubMed]
- Schulz, B.; Boyle, C. What Are Endophytes? In Microbial Root Endophytes; Schulz, B.J.E., Boyle, C.J.C., Sieber, T.N., Eds.; Springer: Berlin, Germany, 2006; pp. 1–13. [Google Scholar] [CrossRef]
- Latch, G.C.M. Physiological Interactions of Endophytic Fungi and Their Hosts. Biotic Stress Tolerance Imparted to Grasses by Endophytes. Agric. Ecosyst. Environ. 1993, 44, 143–156. [Google Scholar] [CrossRef]
- Huitu, O.; Forbes, K.M.; Helander, M.; Julkunen-Tiitto, R.; Lambin, X.; Saikkonen, K.; Stuart, P.; Sulkama, S.; Hartley, S. Silicon, Endophytes and Secondary Metabolites as Grass Defenses against Mammalian Herbivores. Front. Plant Sci. 2014, 5, 478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herre, E.A.; Mejía, L.C.; Kyllo, D.A.; Rojas, E.; Maynard, Z.; Butler, A.; Van Bael, S.A. Ecological Implications of Anti-pathogen Effects of Tropical Fungal Endophytes and Mycorrhizae. Ecology 2007, 88, 550–558. [Google Scholar] [CrossRef] [Green Version]
- Schulz, B.; Römmert, A.-K.; Dammann, U.; Aust, H.-J.; Strack, D. The Endophyte-Host Interaction: A Balanced Antagonism? Mycol. Res. 1999, 103, 1275–1283. [Google Scholar] [CrossRef]
- Wilson, D. Endophyte: The Evolution of a Term, and Clarification of Its Use and Definition. Oikos 1995, 73, 274–276. [Google Scholar] [CrossRef]
- Freeman, S.; Horowitz, S.; Sharon, A. Pathogenic and Nonpathogenic Lifestyles in Colletotrichum Acutatum from Strawberry and Other Plants. Phytopathology 2001, 91, 986–992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maciá-Vicente, J.G.; Piepenbring, M.; Koukol, O. Brassicaceous Roots as an Unexpected Diversity Hot-Spot of Helotialean Endophytes. IMA Fungus 2020, 11, 16. [Google Scholar] [CrossRef] [PubMed]
- Spadaro, D.; Galliano, A.; Pellegrino, C.; Gilardi, G.; Garibaldi, A.; Gullino, M.L. Dry matter and mineral composition, together with commercial storage practices, influence the development of skin pitting caused by Cadophora luteo-olivacea on kiwifruit ‘Hayward’. J. Plant Pathol. 2010, 92, 339–346. [Google Scholar]
- Nakaune, R.; Tatsuki, M.; Matsumoto, H.; Ikoma, Y. First Report of a New Postharvest Disease of Grape Caused by Cadophora Luteo-Olivacea. J. Gen. Plant Path. 2016, 82, 116–119. [Google Scholar] [CrossRef]
- Wenneker, M.; Pham, K.T.K.; Lemmers, M.E.C.; de Boer, F.A.; van Leeuwen, P.J.; Hollinger, T.C.; Groenenboom-de Haas, B.H.; Köhl, J. First Report of Cadophora Luteo-Olivacea Causing Side Rot on ‘Conference’ Pears in the Netherlands. Plant Dis. 2016, 100, 2162. [Google Scholar] [CrossRef]
- Frisullo, S. First report of “Cadophora malorum” on “Asparagus officinalis” in Italy. Phytopathol. Mediter. 2002, 2, 1–4. [Google Scholar] [CrossRef]
- Di Marco, S.; Calzarano, F.; Osti, F.; Mazzullo, A. Pathogenicity of Fungi Associated with a Decay of Kiwifruit. Australas. Plant Pathol. 2004, 33, 337–342. [Google Scholar] [CrossRef]
- Kageyama, S.A.; Mandyam, K.G.; Jumpponen, A. Diversity, Function and Potential Applications of the Root-Associated Endophytes. In Mycorrhiza: State of the Art, Genetics and Molecular Biology, Eco-Function, Biotechnology, Eco-Physiology, Structure and Systematics; Varma, A., Ed.; Springer: Berlin, Germany, 2008; pp. 29–57. [Google Scholar] [CrossRef]
- Köhl, J.; Groenenboom-de Haas, B.; Goossen-van de Geijn, H.; Speksnijder, A.; Kastelein, P.; de Hoog, S.; Gerrits van den Ende, B. Pathogenicity of Stemphylium Vesicarium from Different Hosts Causing Brown Spot in Pear. Eur. J. Plant Pathol. 2008, 124, 151. [Google Scholar] [CrossRef]
- Amadioha, A.C. Control of Storage Rot of Potato Caused by Rhizopus Oryzae. Int. J. Pest Manag. 1996, 42, 311–314. [Google Scholar] [CrossRef]
- Gnanesh, B.N.; Tejaswi, A.; Arunakumar, G.S.; Supriya, M.; Manojkumar, H.B.; Tewary, P. Molecular Phylogeny, Identification and Pathogenicity of Rhizopus Oryzae Associated with Root Rot of Mulberry in India. J. Appl. Microbiol. 2020, 131, 360–374. [Google Scholar] [CrossRef] [PubMed]
- Zajc, J.; Černoša, A.; Di Francesco, A.; Castoria, R.; De Curtis, F.; Lima, G.; Badri, H.; Jijakli, H.; Ippolito, A.; GostinČar, C.; et al. Characterization of Aureobasidium pullulans isolates selected as biocontrol agents against fruit decay pathogens. Fungal Genom. Biol. 2020, 10, 1–13. [Google Scholar] [CrossRef]



| Sampling Site | Locality | Latitude | Longitude | Altitude (m) | Sampled Tissues * | No. of Samples | Life Stage ** |
|---|---|---|---|---|---|---|---|
| 1 | S. Martino in Colle (Umbria) | 43.0335354 | 12.3644377 | 275 | corm | 7 | v |
| leaf | 7 | v | |||||
| tepal | 3 | f | |||||
| stigma | 3 | f | |||||
| 2 | Città della Pieve (Umbria) | 42.9527338 | 12.004326 | 513 | leaf | 5 | v |
| 3 | Moiano (Umbria) | 43.0148483 | 12.0184455 | 268 | corm | 6 | v |
| corm | 5 | f | |||||
| corm * | 6 | f | |||||
| leaf | 6 | v | |||||
| leaf * | 6 | f | |||||
| stem | 8 | f | |||||
| stem * | 6 | f | |||||
| tepal | 3 | f | |||||
| stigma | 3 | f | |||||
| 4 | Gualdo Cattaneo (Umbria) | 42.9094087 | 12.5558159 | 461 | corm | 5 | f |
| tepal | 3 | f | |||||
| 5 | Giano dell’Umbria (Umbria) | 42.8334672 | 12.5777111 | 542 | corm | 4 | v |
| corm | 5 | d | |||||
| corm | 3 | f | |||||
| tepal | 3 | f | |||||
| stigma | 3 | f | |||||
| 6 | Castel Ritaldi (Umbria) | 42.8232601 | 12.6722871 | 297 | tepal | 3 | f |
| 7 | Foligno (Umbria) | 42.9561825 | 12.703334 | 243 | corm | 5 | f |
| 8 | Cantalupo di Bevagna (Umbria) | 42.9683821 | 12.5796837 | 201 | corm | 4 | v |
| corm | 5 | d | |||||
| corm | 3 | f | |||||
| tepal | 3 | f | |||||
| stigma | 3 | f | |||||
| 9 | Città di Castello (Umbria) | 43.4566183 | 12.3247772 | 566 | corm | 2 | d |
| 10 | Zafferana Etnea (Sicily) | 37.6932846 | 15.1064599 | 584 | corm | 2 | v |
| OTU | Closest Match in GenBank | No. of Isolates | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Name | Accession No. | Taxon | Accession No. | % of Similarity | Corms | Stems | Leaves | Tepals | Stigmas | Total |
| ASCOMYCOTA | ||||||||||
| Helotiales | ||||||||||
| 1 | MW798781 | Cadophora luteo-olivacea | HM116747 | 99.8 | 32 | 32 | ||||
| 2 | MW798782 | Cadophora malorum | KF646089 | 99.8 | 19 | 19 | ||||
| 3 | MW798777 | Botrytis cinerea | MH860108 | 100 | 1 | 1 | ||||
| Pleosporales | ||||||||||
| 4 | MW798757 | Alternaria alternata | MT453271 | 100 | 1 | 1 | 13 | 4 | 19 | |
| 5 | MW798753 | Alternaria infectoria | MK461063 | 99.8 | 1 | 1 | ||||
| 6 | MW798751 | Stemphylium vesicarium | MK461018 | 100 | 4 | 4 | ||||
| 7 | MW798765 | Pyrenophora tritici-repentis | MH399396 | 99.8 | 1 | 1 | ||||
| 8 | MW798750 | Epicoccum sp. | HQ630972 | 100 | 7 | 7 | ||||
| 36 | MW798766 | Epicoccum nigrum | MH931271 | 98.4 | (1) | (1) | ||||
| 9 | MW798760 | Stagonosporopsis cucurbitacearum | KM489071 | 98.6 | 1 | 1 | ||||
| 10 | MW798754 | Spegazzinia sp. | KR093917 | 99.1 | 1 | 1 | ||||
| Dothideales | ||||||||||
| 11 | MW798756 | Aureobasidium pullulans | FN868454 | 99.2 | (1) | 2 | 1 | 3 (1) | ||
| 12 | MW798769 | Aureobasidium pullulans | MT153709 | 95.64 | 1 | 1 | ||||
| Capnodiales | ||||||||||
| 13 | MW798749 | Cladosporium cladosporioides | MH863979 | 100 | 1 | 1 | ||||
| Eurotiales | ||||||||||
| 14 | MW798764 | Talaromyces pinophilus | KC867288 | 99.7 | 7 (2) | 2 | 9 (2) | |||
| 15 | MW798762 | Talaromyces cecidicola | MH862736 | 99.8 | 5 | 1 | 6 | |||
| 16 | MW798752 | Talaromyces assiutensis | JN899320 | 99.5 | 1 | 1 | 2 | |||
| 17 | MW798763 | Aspergillus niger | MF422165 | 100 | 2 | 2 | ||||
| 18 | MW798758 | Aspergillus sp. | MK461022 | 100 | 1 | 1 | ||||
| 19 | MW798755 | Aspergillus flavipes | HM595494 | 99.4 | 1 | 1 | ||||
| 20 | MW798761 | Aspergillus europaeus | LT220221 | 99.7 | 1 | 1 | ||||
| 21 | MW798759 | Penicillium citrinum | KX958075 | 99.8 | 1 | 1 | ||||
| Hypocreales | ||||||||||
| 22 | MW798779 | Fusarium oxysporum | MT453296 | 100 | 1 (9) | (3) | (1) | 1 | 2 (13) | |
| 23 | MW798774 | Ilyonectria sp. | KT268970 | 100 | 1 | 1 | ||||
| 24 | MW798780 | Parengyodontium album | LC092887 | 99.6 | 1 | 1 | ||||
| 35 | MW798772 | Trichoderma sp. | MW450867 | 99,8 | (1) | (1) | ||||
| Xylariales | ||||||||||
| 25 | MW798776 | Hypoxylon fuscum | MW367856 | 99.6 | 1 | 1 | ||||
| Sordariales | ||||||||||
| 26 | MW798778 | Ovatospora brasiliensis | MH858514 | 99.8 | 1 | 1 | ||||
| Leotiomycetes_incertae_sedis | ||||||||||
| 27 | MW798775 | Malbranchea circinata | MN627784 | 99.4 | 1 | 1 | ||||
| Saccharomycetales | ||||||||||
| 28 | Meyerozyma caribbica | KY104217 | 100 | 1 | 1 | 2 | ||||
| BASIDIOMYCOTA | ||||||||||
| Sporidiobolales | ||||||||||
| 29 | MW798748 | Rhodotorula sp. | HG936596 | 99.7 | 1 | 1 | 1 | 3 | ||
| Agaricales | ||||||||||
| 30 | MW798746 | Coprinellus micaceus | FN386285 | 99.8 | 1 | 1 | ||||
| Russulales | ||||||||||
| 31 | MW798745 | Peniophora sp. | MT156128 | 99.5 | 1 | 1 | ||||
| Filobasidiales | ||||||||||
| 32 | MW798744 | Filobasidium wieringae | KY103450 | 100 | 1 | 1 | ||||
| 34 | MW798747 | uncultured fungus/Filobasidium sp. | AF444450 | 99.7 | 1 | 1 | ||||
| MUCOROMYCOTA | ||||||||||
| Mucorales | ||||||||||
| 33 | MW798768 | Mucor fragilis | KU319073 | 100 | 2 | 3 | 5 | |||
| 37 | MW798770 | Mucor circinelloides | KP132468 | 99.8 | (1) | (1) | ||||
| 38 | MW798773 | Rhizopus oryzae | MF685318 | 99.9 | (5) | (2) | (7) | |||
| 39 | MW798771 | Rhizopus oryzae | HQ435056 | 98 | (3) | (1) | (4) | |||
| Total No. isolates | 71 (20) | 10 (6) | 19 (4) | 22 | 13 | 135 (30) | ||||
| Species richness | 11 (5) | 9 (3) | 10 (4) | 8 | 8 | 34 (8) | ||||
| OTU | Taxon | Pi | ||||
|---|---|---|---|---|---|---|
| Corms | Stems | Leaves | Tepals | Stigmas | ||
| 1 | Cadophora luteo-olivacea | 0.451 * | ||||
| 2 | Cadophora malorum | 0.268 * | ||||
| 3 | Botrytis cinerea | 0.053 | ||||
| 4 | Alternaria alternata | 0.1 | 0.053 | 0.591 * | 0.308 * | |
| 5 | Alternaria infectoria | 0.045 | ||||
| 6 | Stemphylium vesicarium | 0.211 * | ||||
| 7 | Pyrenophora tritici-repentis | 0.045 | ||||
| 8 | Epicoccum sp. | 0.368 * | ||||
| 9 | Stagonosporopsis cucurbitacearum | 0.045 | ||||
| 10 | Spegazzinia sp. | 0.077 | ||||
| 11 | Aureobasidium pullulans | 0.091 | 0.077 | |||
| 12 | Aureobasidium pullulans | 0.1 | ||||
| 13 | Cladosporium cladosporioides | 0.053 | ||||
| 14 | Talaromyces pinophilus | 0.099 * | 0.2 * | |||
| 15 | Talaromyces cecidicola | 0.070 | 0.1 | |||
| 16 | Talaromyces assiutensis | 0.014 | 0.053 | |||
| 17 | Aspergillus niger | 0.028 | ||||
| 18 | Aspergillus sp. | 0.077 | ||||
| 19 | Aspergillus flavipes | 0.077 | ||||
| 20 | Aspergillus europaeus | 0.1 | ||||
| 21 | Penicillium citrinum | 0.1 | ||||
| 22 | Fusarium oxysporum | 0.014 | 0.077 | |||
| 23 | Ilyonectria sp. | 0.014 | ||||
| 24 | Parengyodontium album | 0.014 | ||||
| 25 | Hypoxylon fuscum | 0.053 | ||||
| 26 | Ovatospora brasiliensis | 0.077 | ||||
| 27 | Malbranchea circinata | 0.053 | ||||
| 28 | Meyerozyma caribbica | 0.014 | 0.1 | |||
| 29 | Rhodotorula sp. | 0.014 | 0.1 | 0.045 | ||
| 30 | Coprinellus micaceus | 0.053 | ||||
| 31 | Peniophora sp. | 0.1 | ||||
| 32 | Filobasidium wieringae | 0.045 | ||||
| 34 | Uncultured fungus/Filobasidium sp. | 0.053 | ||||
| 33 | Mucor fragilis | 0.091 | 0.231 * | |||
| 1/S | 0.091 | 0.111 | 0.1 | 0.125 | 0.125 | |
| OTU | Taxon | Life Stage | ||
|---|---|---|---|---|
| Vegetative | Dormant | Flowering | ||
| 1 | Cadophora luteo-olivacea | 12 | 6 | 14 |
| 2 | Cadophora malorum | 12 | 4 | 3 |
| 14 | Talaromyces pinophilus | 3 | 4 | |
| 15 | Talaromyces cecidicola | 5 | ||
| 16 | Talaromyces assiutensis | 1 | ||
| 17 | Aspergillus sp. | 2 | ||
| 22 | Fusarium oxysporum | 1 | ||
| 23 | Ilyonectria sp. | 1 | ||
| 24 | Parengyodontium album | 1 | ||
| 28 | Meyerozyma caribbica | 1 | ||
| 29 | Rhodotorula sp. | 1 | ||
| Total No. isolates | 30 | 10 | 31 | |
| Species richness | 6 | 2 | 8 | |
| OTU | Taxon | Sampling Site | |||
|---|---|---|---|---|---|
| Bevagna | Moiano | S. Martino in Colle | Giano dell’Umbria | ||
| 1 | Cadophora luteo-olivacea | 11 | 4 | 6 | |
| 2 | Cadophora malorum | 4 | 2 | 2 | |
| 3 | Botrytis cinerea | 1 | |||
| 4 | Alternaria alternata | 9 | 2 | 1 | 4 |
| 5 | Alternaria infectoria | 1 | |||
| 6 | Stemphylium vesicarium | 1 | 2 | ||
| 8 | Epicoccum sp. | 6 | |||
| 9 | Stagonosporopsis cucurbitacearum | 1 | |||
| 10 | Spegazzinia sp. | 1 | |||
| 11 | Aureobasidium pullulans | 3 | |||
| 12 | Aureobasidium pullulans | 1 | |||
| 13 | Cladosporium cladosporioides | 1 | |||
| 14 | Talaromyces pinophilus | 2 | 3 | 3 | |
| 15 | Talaromyces cecidicola | 4 | |||
| 16 | Talaromyces assiutensis | 2 | |||
| 17 | Aspergillus niger | 2 | |||
| 18 | Aspergillus sp. | 1 | |||
| 19 | Aspergillus flavipes | 1 | |||
| 20 | Aspergillus europaeus | 1 | |||
| 21 | Penicillium citrinum | 1 | |||
| 22 | Fusarium oxysporum | 1 | 1 | ||
| 23 | Ilyonectria sp. | 1 | |||
| 24 | Parengyodontium album | 1 | |||
| 26 | Ovatospora brasiliensis | 1 | |||
| 27 | Malbranchea circinata | 1 | |||
| 28 | Meyerozyma caribbica | 2 | |||
| 29 | Rhodotorula sp. | 1 | 1 | 1 | |
| 30 | Coprinellus micaceus | 1 | |||
| 31 | Peniophora sp. | 1 | |||
| 32 | Filobasidium wieringae | 1 | |||
| 33 | Mucor fragilis | 5 | |||
| Total No. isolates | 37 | 36 | 13 | 16 | |
| Species richness | 10 | 20 | 9 | 5 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belfiori, B.; Rubini, A.; Riccioni, C. Diversity of Endophytic and Pathogenic Fungi of Saffron (Crocus sativus) Plants from Cultivation Sites in Italy. Diversity 2021, 13, 535. https://doi.org/10.3390/d13110535
Belfiori B, Rubini A, Riccioni C. Diversity of Endophytic and Pathogenic Fungi of Saffron (Crocus sativus) Plants from Cultivation Sites in Italy. Diversity. 2021; 13(11):535. https://doi.org/10.3390/d13110535
Chicago/Turabian StyleBelfiori, Beatrice, Andrea Rubini, and Claudia Riccioni. 2021. "Diversity of Endophytic and Pathogenic Fungi of Saffron (Crocus sativus) Plants from Cultivation Sites in Italy" Diversity 13, no. 11: 535. https://doi.org/10.3390/d13110535
APA StyleBelfiori, B., Rubini, A., & Riccioni, C. (2021). Diversity of Endophytic and Pathogenic Fungi of Saffron (Crocus sativus) Plants from Cultivation Sites in Italy. Diversity, 13(11), 535. https://doi.org/10.3390/d13110535