Bacterial Community Is Affected by Locations and Time Rather Than Potato Varieties but Streptomyces spp. Are Related to Potato Varieties
Abstract
:1. Introduction
2. Materials and Methods
2.1. Field Site Description
2.2. Soil Sampling and Harvesting of Potato
2.3. Soil Chemical Properties Determination
2.4. Soil DNA Extraction, PCR Amplification and Gene Expression
2.5. Sequencing Data and Diversity Analysis
2.6. Disease Severity Analysis
2.7. Statistical Analysis
3. Results
3.1. Environmental Parameters and Soil Chemical Properties
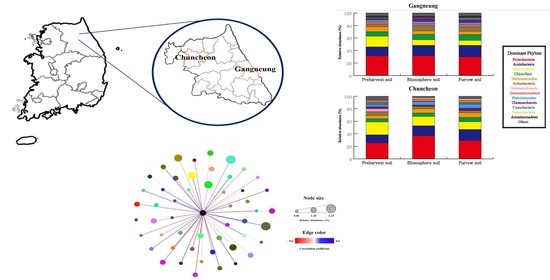
3.2. Composition of Dominant Bacterial Community and Their Similarities
3.3. Bacterial Diversity
3.4. Relative Abundance of Streptomyces spp. in Soil and Its Severity in Potato Varieties
3.5. Correlation between Streptomyces spp. and Other Bacterial Communities
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. Food and Agriculture Organization of the United Nations. Statistic Division. Available online: http://www.fao.org/faostat/en/#data/QC/visualize (accessed on 25 September 2021).
- Zhang, H.; Gao, Z.; Shi, A.; Fang, S. Soil bacterial diversity and its relationship with soil CO2 and mineral composition: A case study of the Laieu experimental site. Int. J. Environ. Res. Public Health 2020, 17, 5699. [Google Scholar] [CrossRef]
- Correa de Souza, R.S.; Armanhi, J.S.L.; Arruda, P. From microbiome to traits: Designing synthetic microbial communities for improved crop resiliency. Front. Plant Sci. 2020, 11, 1179. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, J.; Wei, Y.; Liu, G.; Zeng, H.; Shi, H. Microbiome-wide association studies reveal correlations between the structure and metabolism of the rhizosphere microbiome and disease resistance in cassava. Plant Biotechnol. J. 2021, 19, 689–701. [Google Scholar] [CrossRef]
- Wei, Z.; Gu, Y.; Friman, V.P.; Kowalchuk, G.A.; Xu, Y.; Shen, Q.; Jousset, A. Initial soil microbiome composition and functioning predetermine future plant health. Sci. Adv. 2019, 5, eaaw0759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loria, R.; Kers, J.; Joshi, M. Evolution of plant pathogenicity in Streptomyces. Annu. Rev. Phytopathol. 2006, 44, 469–487. [Google Scholar] [CrossRef]
- Hill, J.; Lazarovits, G. A mail survey of growers to estimate potato common scab prevalence and economic loss in Canada. Can. J. Plant Pathol. 2005, 27, 46–52. [Google Scholar] [CrossRef]
- Dees, M.W.; Wanner, L.A. In search of better management of potato common scab. Potato Res. 2012, 55, 249–268. [Google Scholar] [CrossRef]
- Peters, R.D.; Sturz, A.V.; Carter, M.R.; Sanderson, J.B. Influence of crop rotation and conservation tillage practices on the severity of soil-borne potato diseases in temperate humid agriculture. Can. J. Soil Sci. 2004, 84, 397–402. [Google Scholar] [CrossRef]
- Haynes, K.G.; Wanner, L.A.; Thill, C.A.; Bradeen, J.M.; Miller, J.; Novy, R.G.; Vinyard, B. Common scab trials of potato varieties and advanced selections at three US locations. Ann. J. Potato Res. 2010, 87, 261–276. [Google Scholar] [CrossRef]
- Lapwood, D.H.; Wellings, L.W.; Rosser, W.R. The control common scab of potatoes by irrigation. Ann. Appl. Biol. 1970, 66, 397–405. [Google Scholar] [CrossRef]
- Larkin, R.P.; Honeycutt, C.W.; Griffin, T.S.; Olanya, O.M.; Halloran, J.M.; He, Z. Effects of different potato cropping system approaches and water management on soil borne disease and soil microbial communities. Phytopathology 2011, 101, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Scholte, K.; Labruyère, R.E. Netted scab: A new name for an old disease in Europe. Potato Res. 1985, 28, 443–448. [Google Scholar] [CrossRef]
- Lacey, M.J.; Wilson, C.R. Relationship of common scab incidence of potatoes grown in Tasmanian ferrosol soils with pH, exchangeable cations and other chemical properties of those soils. J. Phytopathol. 2001, 149, 679–683. [Google Scholar] [CrossRef]
- Lambert, D.H.; Loria, R. Streptomyces acidiscabies sp. nov. Int. J. Syst. Evol. Microbiol. 1989, 39, 393–396. [Google Scholar] [CrossRef] [Green Version]
- Lindholm, P.; Kortemaa, H.; Kokkola, M.; Haahtela, K.; Salkinoja-Salonen, M.; Valkonen, J.P.T. Streptomyces spp. isolated from potato scab lesions under Nordic conditions in Finland. Plant Dis. 1997, 81, 1317–1322. [Google Scholar] [CrossRef] [Green Version]
- Al-Mughrabi, K.I.; Vikram, A.; Poirier, R.; Jayasuriya, K.; Moreau, G. Management of common scab of potato in the field using biopesticides, fungicides, soil additives, or soil fumigants. Biocontrol Sci. Technol. 2016, 26, 125–135. [Google Scholar] [CrossRef]
- Larkin, R.P.; Griffin, T.S.; Honeycutt, C.W. Rotation and cover crop effects on soil borne potato disease, tuber yield, and soil microbial communities. Plant Dis. 2010, 94, 1491–1502. [Google Scholar] [CrossRef] [Green Version]
- Wiggins, B.E.; Kinkel, L.L. Green manures and crop sequences influence alfalfa root rot and pathogen inhibitory activity among soil-borne streptomycetes. Plant Soil 2005, 268, 271–283. [Google Scholar] [CrossRef]
- Hiltunen, L.H.; Weckman, A.; Ylhäinen, A.; Rita, H.; Richter, E.; Valkonen, J.P.T. Responses of potato cultivars to the common scab pathogens, Streptomyces scabies and S. turgidiscabies. Ann. Appl. Biol. 2005, 146, 395–403. [Google Scholar] [CrossRef]
- St-Onge, R.; Gadkar, V.J.; Arseneault, T.; Goyer, C.; Filion, M. The ability of Pseudomonas sp. LBUM 223 to produce phenazine-1-carboxylic acid affects the growth of Streptomyces scabies, the expression of thaxtomin biosynthesis genes and the biological control potential against common scab of potato. FEMS Microbial. Ecol. 2011, 75, 173–183. [Google Scholar] [CrossRef] [Green Version]
- Chin, A.; Woeng, T.F.C.; Bloemberg, G.V.; van der Drift, K.M.G.M.; Schripsema, J.; Kroon, B.; Keel, C.; Balser, P.A.H.M.; Tichy, H.V.; de Brujin, F.K.; et al. Biocontrol by phenazine-1-carboxamide-producing Pseudomonas chlorophis PCL1391 of tomato root caused by Fusarium oxysporum f.sp. radicis-lycopersici. Mol. Plant.-Microbe Interact. 1998, 11, 1069–1077. [Google Scholar] [CrossRef] [Green Version]
- Shi, W.; Li, M.; Wei, G.; Tian, R.; Li, C.; Wang, B.; Gao, Z. The occurrence of potato common scab correlates with the community composition and function of the geocaulosphere soil microbiome. Microbiome 2019, 7, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Zhang, H.; Sun, L.; Qi, G.; Chen, S.; Zhao, X. Microbial community composition is related to soil biological and chemical properties and bacterial wilt outbreak. Sci. Rep. 2017, 7, 1–10. [Google Scholar]
- Mendes, R.; Kruijt, M.; De Bruijn, I.; Dekkers, E.; van der Voort, M.; Schneider, J.H.; Raaijmakers, J.M. Deciphering the rhizosphere microbiome for disease suppressive bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef]
- Romaniuk, R.; Giuffre, L.; Costantini, A.; Nannipiere, P. Assessment of soil microbial diversity measurements as indicators of soil functioning in organic and conventional horticulture systems. Ecol. Indic. 2011, 11, 1345–1353. [Google Scholar] [CrossRef]
- Bastida, F.; Torres, I.F.; Moreno, J.L.; Baldrian, P.; Ondono, S.; Ruiz-Navarro, A.; Jehmlich, N. The active microbial diversity drives ecosystem multifunctionality and is physiologically related to carbon availability in Mediterranean semi-arid soils. Mol. Ecol. 2016, 25, 4660–4673. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Jamdagni, P.; Goyal, K. Recent trends in characterization of microbial Diversity from Environment. Acta Agric. Serbica 2012, 17, 31–46. [Google Scholar]
- Lara-Victoriano, F.; Castillo-Reyes, F.; Flores-Gallegos, C.; Aguilar, C.N.; Rodriguez-Herrera, R. Metagenomics in plant pathology. In Phytopathology in the Omics Era; Research Signpost: Trivandrum, India, 2011; ISBN 978-81-308-0438-5. [Google Scholar]
- Piombo, E.; Abdelfattah, A.; Droby, S.; Wisniewski, M.; Spadaro, D.; Schena, L. Metagenomics approaches for the detection and surveillance of emerging and recurrent plant pathogens. Microorganisms 2021, 9, 188. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bates, S.T.; Clemente, J.C.; Flores, G.E.; Walters, W.A.; Parfrey, L.W.; Knight, R.; Fierer, N. Global biogeography of highly diverse protistan communities in soil. ISME J. 2013, 7, 652–659. [Google Scholar] [CrossRef]
- Igalavithana, A.D.; Lee, S.S.; Niazi, N.K.; Lee, Y.H.; Kim, K.H.; Park, J.H.; Moon, D.H.; Ok, Y.S. Assessment of soil health in urban agriculture: Soil enzymes and microbial properties. Sustainability 2017, 9, 310. [Google Scholar] [CrossRef] [Green Version]
- Bates, S.T.; Berg-Lyons, D.; Caporaso, J.G.; Walters, W.A.; Knight, R.; Fierer, N. Examining the global distribution of dominant archael populations in soil. ISME J. 2011, 5, 908–917. [Google Scholar] [CrossRef] [Green Version]
- Sagova-Mareckova, M.; Daniel, O.; Omelka, M.; Kristufek, V.; Divis, J.; Kopeck, J. Determination of factors associated with natural soil suppressivity to potato common scab. PLoS ONE 2015, 10, e0116291. [Google Scholar]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. Peer J. 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Song, Z.; Yang, X.; Mao, Z.; Nie, X.; Guo, H.; Peng, X. Microbial community analysis of apple rhizosphere around Bohai Gulf. Sci. Rep. 2017, 7, 8918. [Google Scholar] [CrossRef] [PubMed]
- Sokal, R.R.; Michener, C.D. A statistical method for evaluating systematic relationships. Univ. Kans. Sci. Bull. 1958, 38, 1409–1438. [Google Scholar]
- Van Elsas, J.D.; Chiurazzi, M.; Mallon, C.A.; Elhottova, D.; Kristufek, V.; Salles, J.F. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc. Natl. Acad. Sci. USA 2012, 109, 1159–1164. [Google Scholar] [CrossRef] [Green Version]
- Berg, G.; Smalla, K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbial. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Nahar, K.; Floc’h, J.B.; Goyer, C.; Zebarth, B.J.; Whitney, S. Diversity of Soil Bacterial Community Is Influenced by Spatial Location and Time but Not Potato Cultivar. Phytobiomes J. 2020, 4, 225–238. [Google Scholar] [CrossRef]
- Inceiglu, O.; Ai-Soud, W.A.; Salles, J.F.; Semenov, A.V.; van Elsas, J.D. Comparative analysis of bacterial communities in a potato field as determined by pyrosequencing. PLoS ONE 2011, 6, e23321. [Google Scholar]
- Liu, R.; Pan, Y.; Bao, H.; Liang, S.; Jiang, Y.; Yu, H.; Nong, J.; Huang, W. Variations in soil physico-chemical properties along slope position gradient in secondary vegetation of the hilly region, Guilin, Southwest China. Sustainability 2020, 12, 1303. [Google Scholar] [CrossRef] [Green Version]
- Yimer, F.; Ledin, S.; Abdelkadir, A. Soil organic carbon and total nitrogen stock as affected by topographic aspect and vegetation in the Bale Mountains, Ethiopia. Geoderma 2006, 135, 335–344. [Google Scholar] [CrossRef]
- Hartmann, A.; Schmid, M.; Van Tuinen, D.; Berg, G. Plant-driven selection of microbes. Plant Soil. 2009, 321, 235–257. [Google Scholar] [CrossRef]
- Essarioui, A.; LeBlanc, N.; Kistler, H.C.; Kinkel, L.L. Plant community richness mediates inhibitory interactions and resource competition between Streptomyces and Fusarium populations in the rhizosphere. Microb. Ecol. 2017, 74, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Arseneault, T.; Goyer, C.; Filion, M. Pseudomonas fluorescens LBUM223 increases potato yield and reduces common scab symptoms in the field. Phytopathology 2015, 105, 1311–1317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marschner, P.; Yang, C.H.; Lieberei, R.; Crowley, D.E. Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol. Biochem. 2001, 33, 1437–1445. [Google Scholar] [CrossRef]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef] [Green Version]
- Waldrop, M.P.; Zak, D.R.; Blackwood, C.B.; Curtis, C.D.; Tilman, D. Resource availability controls fungal diversity across a plant diversity gradient. Ecol. Lett. 2006, 10, 1127–1135. [Google Scholar] [CrossRef] [Green Version]
- Delgado-Baquerizo, M.; Reich, P.B.; Khachane, A.N.; Campbell, C.D.; Thomas, N.; Freitag, T.E.; Singh, B.K. It is elemental: Soil nutrient stoichiometry drives bacterial diversity. Environ. Microbiol. 2017, 19, 1176–1188. [Google Scholar] [CrossRef]
- Zhou, J.; Deng, Y.; Shen, L.; Wen, C.; Yan, Q.; Ning, D.; Voordeckers, J.W. Temperature mediates continental-scale diversity of microbes in forest soils. Nat. Commun. 2016, 7, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Maestre, F.T.; Delgado-Baquerizo, M.; Jeffries, T.C.; Eldridge, D.J.; Ochoa, V.; Gozalo, B.; Bowker, M.A. Increasing aridity reduces soil microbial diversity and abundance in global drylands. Proc. Natl. Acad. Sci. USA 2015, 112, 15684–15689. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Field Sites | Harvest Time (Spatial Location) | pH | EC (dsm−1) | SOM (gkg−1) | Ca2+ | K+ | Mg2+ | Na+ |
|---|---|---|---|---|---|---|---|---|
| (Cmolkg−1) | ||||||||
| Gangneung | Preharvest | 6.05 ± 0.27 ab | 1.13 ± 0.43 a | 11.15 ± 0.82 a | 7.74 ± 0.18 a | 0.18 ± 0.03 a | 2.54 ± 0.13 a | 0.16 ± 0.01 a |
| Postharvest (Rhizosphere) | 5.64 ± 0.09 a | 0.59 ± 0.13 ab | 13.94 ± 1 b | 5.68 ± 0.37 b | 0.12 ± 0.02 a | 0.6 ± 0.03 b | 0.19 ± 0.01 a | |
| Postharvest (Furrow) | 6.37 ± 0.21 a | 0.35 ± 0.02 b | 14.83 ± 2.37 a | 5.66 ± 0.14 b | 0.1 ± 0.01 b | 1.61 ± 0.07 b | 0.17 ± 0.02 a | |
| Chuncheon | Preharvest | 6.43 ± 0.28 b | 4.23 ± 1.56 a | 19 ± 1.34 a | 5.52 ± 1.03 a | 1.12 ± 0.77 a | 1.35 ± 0.24 a | 0.54 ± 0.41 a |
| Postharvest (Rhizosphere) | 7.36 ± 0.04 a | 0.53±0.06 a | 25.72 ± 5.47 a | 3.83 ± 0.34 ab | 0.11 ± 0.02 a | 0.83 ± 0.03 a | 0.16 ± 0.01 a | |
| Postharvest (Furrow) | 7.23 ± 0.06 a | 0.34±0.03 a | 24.99 ± 2.9 a | 3.18 ± 0.24 b | 0.11 ± 0.01 a | 0.81 ± 0.01 b | 0.15 ± 0.01 a | |
| Order/Genus | R-Value (p < 0.05) | Order/Genus | R-Value (p < 0.05) |
|---|---|---|---|
| Bryobacter | 0.495 | Flavobacterium | −0.554 |
| SBR1031/uncultured bacterium | 0.514 | Sphingobacteriales/uncultured bacterium | −0.495 |
| Chloroflexales/Uncultured | 0.492 | Saccharimonadales/uncultured bacterium | −0.562 |
| Tepidisphaerales/Other | 0.471 | Massilla | −0.502 |
| Pseudolabrys | 0.484 | Cellvibrio | −0.667 |
| Sphingomonas | 0.541 | Pseudomonas | 0.576 |
| Other/Other | 0.551 | Chthoniobacter | 0.557 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, G.S.; Adhikari, M.; Yang, J.E.; Kim, H.S.; Han, K.S.; Ha, K.-S.; Park, D.H. Bacterial Community Is Affected by Locations and Time Rather Than Potato Varieties but Streptomyces spp. Are Related to Potato Varieties. Diversity 2021, 13, 659. https://doi.org/10.3390/d13120659
Lee GS, Adhikari M, Yang JE, Kim HS, Han KS, Ha K-S, Park DH. Bacterial Community Is Affected by Locations and Time Rather Than Potato Varieties but Streptomyces spp. Are Related to Potato Varieties. Diversity. 2021; 13(12):659. https://doi.org/10.3390/d13120659
Chicago/Turabian StyleLee, Geon Seung, Mahesh Adhikari, Jae E. Yang, Hyuck Soo Kim, Kyu Suk Han, Kean-Soo Ha, and Duck Hwan Park. 2021. "Bacterial Community Is Affected by Locations and Time Rather Than Potato Varieties but Streptomyces spp. Are Related to Potato Varieties" Diversity 13, no. 12: 659. https://doi.org/10.3390/d13120659
APA StyleLee, G. S., Adhikari, M., Yang, J. E., Kim, H. S., Han, K. S., Ha, K. -S., & Park, D. H. (2021). Bacterial Community Is Affected by Locations and Time Rather Than Potato Varieties but Streptomyces spp. Are Related to Potato Varieties. Diversity, 13(12), 659. https://doi.org/10.3390/d13120659