18S rRNA Analysis Reveals High Diversity of Phytoplankton with Emphasis on a Naked Dinoflagellate Gymnodinium sp. at the Han River (Korea)
Abstract
:1. Introduction
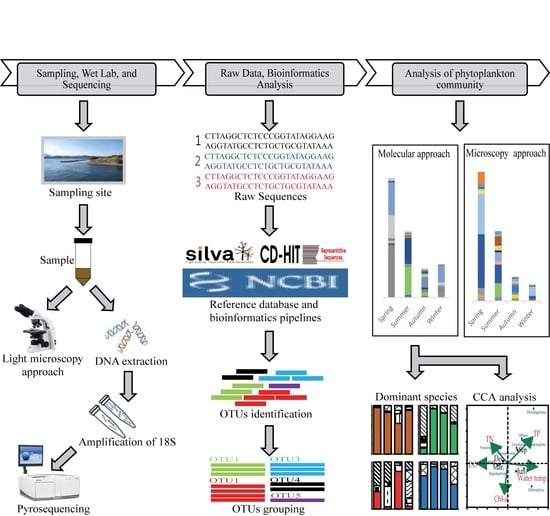
2. Materials and Methods
2.1. Water Sample Collections and Environmental Factors
2.2. Nutrient and Chlorophyll-a Measurement
2.3. Genomic DNA Extraction
2.4. Amplicon PCR and Sequencing
2.5. Sequencing Read Processing, Data Cleaning, and Taxonomic Affiliation
2.6. Diversity Analysis
2.7. Molecular Cloning and Quantitative PCR (qPCR) Detection
3. Results
3.1. Environmental and Biological Factors
3.2. Sequencing Characteristics and Diversity Indices
3.3. Taxonomic Composition of Microeukaryotes
3.4. Seasonal Pattern and Community Composition of Phytoplankton
3.5. Seasonal Changes in the Dominant Species
3.6. 18S rRNA Cloning and Molecular Detection of Gymnodinium sp.
3.7. Comparison of Morphological and Molecular Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Graham, L.E.; Graham, J.M.; Lee, W.W. Algae, 2nd ed.; Benjamin Cummings: San Francisco, CA, USA, 2009. [Google Scholar]
- What Are Phytoplankton? Available online: https://oceanservice.noaa.gov/facts/phyto.html (accessed on 2 June 2020).
- Rakocevic-Nedovic, J.; Hollert, H. Phytoplankton community and chlorophyll a as trophic state indices of Lake Skadar (Montenegro, Balkan). Environ. Sci. Pollut. Res. 2005, 12, 146–152. [Google Scholar] [CrossRef]
- Anderson, D.; Glibert, P.; Burkholder, J. Harmful algal blooms and eutrophication: Nutrient sources, composition, and consequences. Estuaries 2002, 25, 704–726. [Google Scholar] [CrossRef]
- Imhoff, J.F. New dimensions in microbial ecology—Functional genes in studies to unravel the biodiversity and role of functional microbial groups in the environment. Microorganisms 2016, 4, 19. [Google Scholar] [CrossRef]
- Huo, S.; Li, X.; Xi, B.; Zhang, H.; Ma, C.; He, Z. Combining morphological and metabarcoding approaches reveals the freshwater eukaryotic phytoplankton community. Environ. Sci. Eur. 2020, 32, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Siano, R.; Kooistra, W.H.; Montresor, M.; Zingone, A. Unarmoured and thin-walled dinoflagellates from the Gulf of Naples, with the description of Woloszynskia cincta sp. nov. (Dinophyceae, Suessiales). Phycologia 2009, 48, 44–65. [Google Scholar] [CrossRef]
- Gómez, F.; Richlen, M.L.; Anderson, D.M. Molecular characterization and morphology of Cochlodinium strangulatum, the type species of Cochlodinium, and Margalefidinium gen. nov. for C. polykrikoides and allied species (Gymnodiniales, Dinophyceae). Harmful Algae 2017, 63, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Ebenezer, V.; Medlin, L.K.; Ki, J.S. Molecular detection, quantification, and diversity evaluation of microalgae. Mar. Biotech. 2012, 14, 129–142. [Google Scholar] [CrossRef]
- Sun, F.; Pei, H.Y.; Hu, W.R.; Song, M.M. A multi-technique approach for the quantification of Microcystis aeruginosa FACHB-905 biomass during high algae-laden periods. Environ. Technol. 2012, 33, 1773–1779. [Google Scholar] [CrossRef]
- Rohrlack, T.; Edvardsen, B.; Skulberg, R.; Halstvedt, C.B.; Tkilen, H.C. Oligopeptide chemotypes of the toxic freshwater cyanobacterium Planktothrix can form subpopulations with dissimilar ecological traits. Limnol. Oceanogr. 2008, 53, 1279–1293. [Google Scholar] [CrossRef]
- Shang, L.; Hu, Z.; Deng, Y.; Liu, Y.; Zhai, X.; Chai, Z.; Liu, X.; Zhan, Z.; Dobbs, F.C.; Tang, Y.Z. Metagenomic sequencing identifies highly diverse assemblages of dinoflagellate cysts in sediments from ships’ ballast tanks. Microorganisms 2019, 7, 250. [Google Scholar] [CrossRef] [Green Version]
- Eiler, A.; Drakare, S.; Bertilsson, S.; Pernthaler, J.; Peura, S.; Rofner, C.; Simek, K.; Yang, Y.; Znachor, P.; Lindstrãm, E.S.; et al. Unveiling distribution patterns of freshwater phytoplankton by a next generation sequencing based approach. PLoS ONE 2013, 8, e53516. [Google Scholar] [CrossRef]
- Boopathi, T.; Ki, J.S. Unresolved diversity and monthly dynamics of eukaryotic phytoplankton in a temperate freshwater reservoir explored by pyrosequencing. Mar. Freshw. Res. 2016, 67, 1680–1691. [Google Scholar] [CrossRef]
- Jeong, S.W.; Lee, J.H.; Yu, J.S. Environmental studies of the lower part of the Han River V. Blooming characteristics of phytoplankton communities. Algae 2003, 18, 255–262. [Google Scholar] [CrossRef]
- Jung, S.W.; Kwon, O.Y.; Lee, J.H.; Han, M.S. Effects of water temperature and silicate on the winter blooming diatom Stephanodiscus hantzschii (Bacillariophyceae) growing in eutrophic conditions in the lower Han River, South Korea. J. Freshw. Ecol. 2009, 24, 219–226. [Google Scholar] [CrossRef] [Green Version]
- Jung, S.W.; Kwon, O.Y.; Yun, S.M.; Joo, H.M.; Kang, J.-H.; Lee, J.H. Impacts of dam discharge on river environments and phytoplankton communities in a regulated river system, the lower Han River of South Korea. J. Ecol. Environ. 2014, 37, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Parsons, T.R.; Maita, Y.; Lalli, C.M.I. A Manual of Chemical and Biological Methods for Seawater Analysis; Pergamon Press: Oxford, UK, 1984. [Google Scholar]
- Faria, D.; Lee, M.-D.; Lee, J.-B.; Lee, J.; Chang, M.; Youn, S.; Suh, Y.; Ki, J.-S. Molecular diversity of phytoplankton in the East China Sea around Jeju Island (Korea), unraveled by pyrosequencing. J. Oceanogr. 2014, 70, 11–23. [Google Scholar] [CrossRef]
- Ki, J.S. Hypervariable regions (V1–V9) of the dinoflagellate 18S rRNA using a large dataset for marker considerations. J. Appl. Phycol. 2012, 24, 1035–1043. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Li, S.; Chou, H.H. LUCY2: An interactive DNA sequence quality trimming and vector removal tool. Bioinformatics 2004, 20, 2865–2866. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [Green Version]
- Hammer, Ø.; Harper, D.A.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Scott, J.T.; Marcarelli, A. Cyanobacteria in freshwater benthic environments. In Ecology of Cyanobacteria II; Whitton, B.A., Ed.; Springer: Dordrecht, The Netherlands, 2012; pp. 271–289. [Google Scholar]
- Grigorszky, I.; Tihamér Kiss, K.; Pór, G.; Dévai, G.; Nagy, A.S.; Somlyai, I.; Ács, É. Temperature and growth strategies as the essential factors influencing the occurrence of Stephanodiscus minutulus (Kützing) Cleve & Möller and Palatinus apiculatus (Ehrenberg) Craveiro, Calado, Daugbjerg & Moestrup. Fundam. Appl. Limnol. 2017, 189, 167–175. [Google Scholar] [CrossRef]
- Oliveira, S.A.; Ferragut, C.; Bicudo, C.E.M. Relationship between phytoplankton structure and environmental variables in tropical reservoirs with different trophic states. Acta Bot. Bras. 2020, 34, 83–93. [Google Scholar] [CrossRef]
- Brito, A.; Brotas, V.; Caetano, M.; Coutinho, T.P.; Bordalo, A.; Icely, J.; Neto, J.; Serôdio, J.; Moita, T. Defining phytoplankton class boundaries in Portuguese transitional waters: An evaluation of the ecological quality status according to the water framework directive. Ecol. Indic. 2012, 19, 5–15. [Google Scholar] [CrossRef]
- Domingues, R.B.; Barbosa, A.; Galvão, H. Constraints on the use of phytoplankton as a biological quality element within the Water Framework Directive in Portuguese waters. Mar. Pollut. Bull. 2008, 56, 1389–1395. [Google Scholar] [CrossRef] [PubMed]
- Spatharis, S.; Tsirtsis, G. Ecological quality scales based on phytoplankton for the implementation of Water Framework Directive in the Eastern Mediterranean. Ecol. Indic. 2010, 10, 840–847. [Google Scholar] [CrossRef]
- Yu, Z.; Yang, J.; Zhou, J.; Yu, X.; Liu, L.; Lv, H. Water stratification affects the microeukaryotic community in a subtropical deep reservoir. J. Euk. Microbiol. 2014, 61, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Hamady, M.; Knight, R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res. 2009, 19, 1141–1152. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.; Yoon, S.K. A study on the phytoplankton in the Paldang Dam reservoir. III. The changes of diatom community structure. Algae Korean Phycol. Soc. 1996, 11, 277–283. [Google Scholar]
- Ha, K.; Kim, H.W.; Joo, G.J. The phytoplankton succession in the lower part of hypertrophic Nakdong River (Mulgum), South Korea. In Phytoplankton and Trophic Gradients; Springer: Dordrecht, The Netherlands, 1998; pp. 217–227. [Google Scholar]
- Rollo, F.; Sassaroli, S.; Boni, L.; Marota, I. Molecular typing of the red-tide dinoflagellate Gonyaulax polyedra in phytoplankton suspensions. Aquat. Microb. Ecol. 1995, 9, 55–61. [Google Scholar] [CrossRef]
- Rott, E. Some aspects of the seasonal distribution of flagellates in mountain lakes. In Flagellates in Freshwater Ecosystems. Developments in Hydrobiology; Jones, R.I., Ilmavirta, V., Eds.; Springer: Dordrecht, The Netherlands, 1988; Volume 45, pp. 159–170. [Google Scholar]
- Bolch, C.J.; Subramanian, T.A.; Green, D.H. The toxic dinoflagellate Gymnodinium catenatum (Dinophyceae) requires marine bacteria for growth 1. J. Phycol. 2011, 47, 1009–1022. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.J.; Yoo, Y.D.; Kang, N.S.; Rho, J.R. Ecology of Gymnodinium aureolum. I. Feeding in western Korean waters. Aquat. Microbiol. Ecol. 2010, 59, 239–255. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.B.; Kim, G.B. New records of five unarmored genera of the family Gymnodiniaceae (Dinophyceae) in Korean Waters. Korean J. Environ. Biol. 2017, 35, 273–288. [Google Scholar] [CrossRef]
- Kaarunya, E.; Sundari, M.S.; Somasundaram, S.T.; Anantharaman, P. Simple approach combining microscopic and molecular techniques to identify diatoms isolated from Vellar Estuary, South East Coast of India. J. Algal Biomass Util. 2016, 7, 71–77. [Google Scholar]
- Lopes, V.R.; Ramos, V.; Martins, A.; Sousa, M.; Welker, M.; Antunes, A.; Vasconcelos, V.M. Phylogenetic, chemical and morphological diversity of cyanobacteria from Portuguese temperate estuaries. Mar. Environ. Res. 2012, 73, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Tapolczai, K.; Keck, F.; Bouchez, A.; Rimet, F.; Vasselon, V. Diatom DNA metabarcoding for biomonitoring: Strategies to avoid major taxonomical and bioinformatical biases limiting molecular indices capacities. Front. Ecol. Evol. 2019, 7, 409. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Nan, J.; Li, J. Driving factors and dynamics of phytoplankton community and functional groups in an estuary reservoir in the Yangtze River, China. Water 2019, 11, 1184. [Google Scholar] [CrossRef] [Green Version]
- Leland, H.V. The influence of water depth and flow regime on phytoplankton biomass and community structure in a shallow, lowland river. Hydrobiologia 2003, 506, 247–255. [Google Scholar] [CrossRef]






| Month | Total Reads | Classified Reads | Rejected Reads | EP Reads | No. of EP OTU | % of EP |
|---|---|---|---|---|---|---|
| Mar. | 12,587 | 12,046 | 541 | 7579 | 110 | 63 |
| Jun. | 9407 | 9012 | 395 | 5083 | 156 | 56 |
| Sep. | 7138 | 6685 | 453 | 2540 | 174 | 38 |
| Dec. | 4387 | 4181 | 206 | 2506 | 121 | 60 |
| Total | 33,519 | 31,924 | 1595 | 17,708 | 561 | 55.5 |
| Parameter | Month | |||
|---|---|---|---|---|
| Mar. | Jun. | Sep. | Dec. | |
| Shannon (H′) | 2.11 | 2.75 | 3.99 | 2.89 |
| Evenness | 0.08 | 0.10 | 0.30 | 0.16 |
| Chao-1 | 121 | 170 | 207 | 134 |
| Good’s coverage | 95.6 | 98.7 | 99.3 | 98.2 |
| Month | Species Detected through Microscopy | Ratio (%) | Phylotypes Detected through Amplicon Sequencing | Ratio (%) |
|---|---|---|---|---|
| Mar. | Stephanodiscus sp. | 72.8 | Cyclostephanos tholiformis | 38.1 |
| Cyclotella sp. | 5.4. | Stephanodiscus sp. | 24.1 | |
| Cryptomonas sp. | 2.7 | Stephanodiscus hantzschi | 16.7 | |
| Jun. | Stephanodiscus sp. | 26.8 | Chlamydomonas sp. | 30.5 |
| Fragilaria sp. | 12.8 | Cyclotella sp. | 28.1 | |
| Actinastrum sp. | 11.4 | Uncultured Cryptophyte | 2.3 | |
| Sep. | Fragilaria sp. | 13.6 | Gymnodinium sp. | 15.6 |
| Navicula sp. | 9.7 | Chlamydomonas sp. | 7.3 | |
| Stephanodiscus sp. | 5.0 | Cyclotella choctawhatcheeana | 4.4 | |
| Dec. | Stephanodiscus spp. | 68.4 | Stephanodiscus sp. | 36.7 |
| Navicula sp. | 14.1 | Cyclostephanos tholiformis. | 13.2 | |
| Chlamydomonas sp. | 10.9 | Stephanodiscus hantzschi | 5.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muhammad, B.L.; Kim, T.; Ki, J.-S. 18S rRNA Analysis Reveals High Diversity of Phytoplankton with Emphasis on a Naked Dinoflagellate Gymnodinium sp. at the Han River (Korea). Diversity 2021, 13, 73. https://doi.org/10.3390/d13020073
Muhammad BL, Kim T, Ki J-S. 18S rRNA Analysis Reveals High Diversity of Phytoplankton with Emphasis on a Naked Dinoflagellate Gymnodinium sp. at the Han River (Korea). Diversity. 2021; 13(2):73. https://doi.org/10.3390/d13020073
Chicago/Turabian StyleMuhammad, Buhari Lawan, Taehee Kim, and Jang-Seu Ki. 2021. "18S rRNA Analysis Reveals High Diversity of Phytoplankton with Emphasis on a Naked Dinoflagellate Gymnodinium sp. at the Han River (Korea)" Diversity 13, no. 2: 73. https://doi.org/10.3390/d13020073
APA StyleMuhammad, B. L., Kim, T., & Ki, J.-S. (2021). 18S rRNA Analysis Reveals High Diversity of Phytoplankton with Emphasis on a Naked Dinoflagellate Gymnodinium sp. at the Han River (Korea). Diversity, 13(2), 73. https://doi.org/10.3390/d13020073