Molecular Diversity and Evolution of Antimicrobial Peptides in Musca domestica
Abstract
:1. Introduction
2. Materials and Methods
2.1. Database Searches
2.2. Characteristics Identification
2.3. Phylogenetic Tree Construction
2.4. Structure Modeling and Analysis
2.5. Positive Selection Analysis
2.6. Co-Evolutionary Analysis
3. Results
3.1. Cysteine-Containing Peptides
3.1.1. MdDefensins
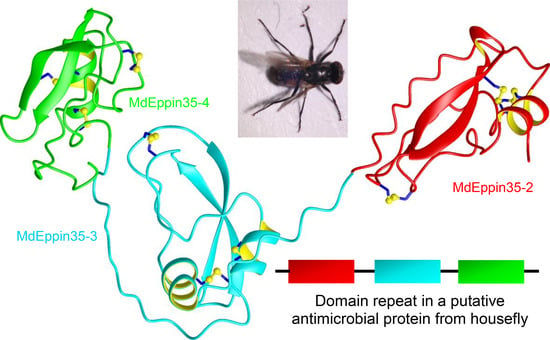
3.1.2. MdEppins
3.1.3. MdMuslins
3.1.4. SVWC Domain AMPs
3.1.5. Crustin-like AMPs
3.2. Linear α-helical Peptides
Cecropins
3.3. Specific Amino Acid-Rich AMPs
3.3.1. Domesticins, Diptericins and Edins
3.3.2. Attacins
4. Discussion
4.1. Gene Duplication
4.2. Exon Duplication via Shuffling
4.3. Protein Terminal Variations
4.4. Evolution of New Disulfide Bridges
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Name | WGS/EST | GenBank No. | Type | Size | MW(Da) | Net Charge | pI | |
|---|---|---|---|---|---|---|---|---|
| ORF | MP | |||||||
| MdDefensin1 # | AIP98387.1 | Csαβ | 93 | 41 | 4158.74 | 3.2 | 8.29 | |
| MdDefensin2 # | AY260152.1 | AAP33451.1 | Csαβ | 92 | 40 | 3995.56 | 3.2 | 8.31 |
| MdDefensin3 # | KJ867444.1 | AIL24687.1 | Csαβ | 99 | 40 | 3995.56 | 3.2 | 8.31 |
| MdDefensin4 # | AQPM01000006.1:16485-16782 | XP_005174767.1 | Csαβ | 93 | 40 | 3995.56 | 3.2 | 8.31 |
| MdDefensin5 # | EF175879.1 | ABM66377.1 | Csαβ | 93 | 40 | 3995.56 | 3.2 | 8.31 |
| MdDefensin6 # | AQPM01000006.1:14681-14958 | XP_005174766.1 | Csαβ | 92 | 40 | 3996.56 | 3.2 | 8.31 |
| MdDefensin7 # | AQPM01000006.1:10815-10994 | AGS57597.1 | Csαβ | 91 | 40 | 4009.59 | 3.2 | 8.31 |
| MdDefensin8 | AQPM01000006.1:19117-19389 | XP_005174768.1 | Csαβ | 91 | 40 | 4228.88 | 4.2 | 8.55 |
| MdDefensin9 | KM047667.1 | Csαβ | 97 | 41 | 4158.74 | 3.2 | 8.29 | |
| MdDefensin10 | AQPM01000006.1:27384-27647 | Csαβ | 88 | 44 | 4642.39 | 3.9 | 8.29 | |
| MdDefensin11 | NDYK01163469.1:1738-2002 | Csαβ | 72 | 44 | 4642.39 | 3.9 | 8.29 | |
| MdDefensin12 | NDYK01012409.1:563-805 | Csαβ | 81 | 41 | 4454.16 | 2.7 | 8.29 | |
| MdDefensin13 # | AQPM01000009.1:1990-2309 | XP_005174769.1 | Csαβ | 83 | 42 | 4383.17 | 2.0 | 8.03 |
| MdDefensin14 | NDYK01067664.1:1236-24 | Csαβ | 83 | 42 | 4383.17 | 2.0 | 8.03 | |
| MdDefensin15 # | AQPM01000008.1:929-1186 | XP_011291282.1 | Csαβ | 86 | 41 | 4232.99 | 2.2 | 8.03 |
| MdDefensin16 # | AQPM01000010.1:7421-7708 | XP_011292449.1 | Csαβ | 96 | 43 | 4707.76 | 6.0 | 8.99 |
| MdDefensin17 | XP_011290793.1 | Csαβ | 75 | 52 | 5492.29 | 1.0 | 7.66 | |
| MdDefensin18 | AQPM01000010.1:5050-5322 | XP_019893215.1 | Csαβ | 91 | 41 | 4495.13 | 3.0 | 8.29 |
| MdDefensin19 | AQPM01000010.1:909-1677 | Csαβ | 84 | 38 | 4360.19 | 2.7 | 8.29 | |
| MdDefensin20 | NDYK01134510.1:970-1187 | Csαβ | 64 | 45 | 5075.93 | 0.9 | 7.61 | |
| MdDefensin21 | AQPM01000007.1:876-1139 | Csαβ | 88 | 65 | 7013.82 | 1.0 | 7.66 | |
| MdEppin1 | XP_005182439.1 | kuntiz domain | 120 | 90 | 10213.55 | −1.2 | 5.55 | |
| MdEppin2 | NDYK01073931.1:1975-2334 | kuntiz domain | 103 | 73 | 8437.57 | 1.7 | 7.98 | |
| MdEppin3 | AQPM01068620.1:8089-8448 | kuntiz domain | 103 | 73 | 8437.57 | 1.7 | 7.98 | |
| MdEppin4 | AQPM01069148.1:21728-22230 | XP_005182685.1 | kuntiz domain | 168 | 149 | 16683,41 | −19 | 4.53 |
| MdEppin5 | XP_005192098.1 | kuntiz domain | 108 | 80 | 9321.66 | 4.7 | 8.71 | |
| MdEppin6 | XP_005192099.1 | kuntiz domain | 201 | 173 | 19487.26 | −1.2 | 5.73 | |
| MdEppin7 | NDYK01008682:1427-1791 | kuntiz domain | 99 | 75 | 8489.55 | 1.2 | 7.66 | |
| MdEppin8 | XP_005182689.1 | kuntiz domain | 93 | 74 | 8636.78 | 7.0 | 9.23 | |
| MdEppin9 | AQPM01082327.1:250-573 | kuntiz domain | 108 | 91 | 10121.48 | −1.1 | 5.97 | |
| MdEppin10 | XP_019893036.1 | kuntiz domain | 83 | 66 | 7318.19 | 1,0 | 7.66 | |
| MdEppin11 | AQPM01068767.1:4133-4939 | kuntiz domain | 83 | 60 | 7255.20 | 7.9 | 9.13 | |
| MdEppin12 | AQPM01058660.1:4502-4813 | kuntiz domain | 104 | 83 | 9255.26 | −2.1 | 5.27 | |
| MdEppin13 | XP_011291014.1 | kuntiz domain | 84 | 63 | 6886.53 | −6.0 | 4.30 | |
| MdEppin14 | AQPM01069148.1:1494-1791 | XP_005182706.1 | kuntiz domain | 85 | 63 | 6862.55 | −3.0 | 4.87 |
| MdEppin15 | AQPM01069148.1:7578-7914 | XP_005182682.2 | kuntiz domain | 91 | 63 | 6966.53 | −8.0 | 4.26 |
| MdEppin16 | AQPM01069148.1:12552-12854 | XP_005182683.1 | kuntiz domain | 82 | 63 | 6946.74 | −2.0 | 5.27 |
| MdEppin17 | AQPM01069148.1:3783-4100 | XP_005182681.1 | kuntiz domain | 84 | 64 | 7101.81 | −2.8 | 5.36 |
| MdEppin18 | AQPM01069148.1:15772-16090 | kuntiz domain | 86 | 66 | 7424.27 | −1.0 | 5.97 | |
| MdEppin19 | XP_005182684.1 | kuntiz domain | 83 | 63 | 7036.78 | −3.0 | 5.05 | |
| MdEppin20 | NDYK01230132:2648-2965 | kuntiz domain | 84 | 64 | 7098.93 | −1.8 | 5.78 | |
| MdEppin21 | NDYK01230132:3440-3753 | kuntiz domain | 87 | 67 | 7507.45 | −1.6 | 6.25 | |
| MdEppin22 | XM_005182623.3 | kuntiz domain | 83 | 64 | 7162.96 | −2.5 | 5.69 | |
| MdEppin23 | XP_005182678.1 | kuntiz domain | 79 | 59 | 6444.22 | −3.0 | 4.94 | |
| MdEppin24 | AQPM01069146.1:10524-10829 | XP_011292137.1 | kuntiz domain | 78 | 59 | 6581.38 | 2.7 | 8.27 |
| MdEppin25 | AQPM01069146.1:7808-8193 | XP_019892012.1 | kuntiz domain | 107 | 88 | 10130.34 | −5.2 | 4.75 |
| MdEppin26 | XP_019892010.1 | kuntiz domain | 78 | 59 | 6468.20 | −3.0 | 4.94 | |
| MdEppin27 | XP_005190040.1 | kuntiz domain | 88 | 68 | 7497.46 | 7.7 | 9.41 | |
| MdEppin28 | NDYK01190170:236-565 | kuntiz domain | 82 | 62 | 7018.89 | 8.7 | 9.70 | |
| MdEppin29 | AQPM01002184.1:2478-2770 | XP_005192097.1 | kuntiz domain | 77 | 57 | 6485.14 | −1.0 | 5.97 |
| MdEppin30 | XP_019895340.1 | kuntiz domain | 88 | 68 | 7688.60 | 4.2 | 8.50 | |
| MdEppin31 | AQPM01011896.1:10591-10914 | kuntiz domain | 85 | 65 | 7374.26 | 4.2 | 8.50 | |
| MdEppin32 | NDYK01025868:2318-2646 | kuntiz domain | 85 | 65 | 7445.39 | 6.2 | 8.95 | |
| MdEppin33 | XP_005182687.1 | kuntiz domain | 145 | 120 | 12984.71 | 7.7 | 9.13 | |
| MdEppin34 | AQPM01069148.1:24856-25351 | XP_019892019.1 | kuntiz domain | 146 | 121 | 13055.78 | 7.7 | 9.13 |
| MdEppin35-1 | NDYK01174947.1:14053-17900 | kuntiz domain | 638 | 46 | 5130.92 | 0.4 | 7.23 | |
| MdEppin35-2 | kuntiz domain | 74 | 8622.47 | −5.0 | 4.80 | |||
| MdEppin35-3 | kuntiz domain | 86 | 9705.43 | −9.0 | 4.46 | |||
| MdEppin35-4 | kuntiz domain | 84 | 9442.15 | −14.2 | 4.05 | |||
| MdEppin35-5 | kuntiz domain | 58 | 6481.97 | −7.0 | 4.23 | |||
| MdEppin35-6 | kuntiz domain | 65 | 7352.07 | −4.8 | 4.77 | |||
| MdEppin35-7 | kuntiz domain | 60 | 6626.16 | −4.0 | 4.70 | |||
| MdEppin35-8 | kuntiz domain | 71 | 8226.17 | −0.3 | 6.44 | |||
| MdEppin35-9 | kuntiz domain | 81 | 9210.26 | 0.7 | 7.61 | |||
| MdMuslin1 | AQPM01019378.1:40908-41220 | kazal domain | 81 | 61 | 6923.73 | −2.7 | 5.36 | |
| MdMuslin2 | AQPM01019378.1:43260-44056 | kazal domain | 75 | 56 | 6374.40 | −0.1 | 6.95 | |
| MdMuslin3 | AQPM01019378.1:47740-48051 | kazal domain | 81 | 62 | 6880.61 | −4.3 | 4.50 | |
| MdMuslin4 | AQPM01019378.1:52156-52447 | kazal domain | 76 | 57 | 6384.33 | −1.0 | 5.88 | |
| MdMuslin5 | AQPM01019378.1:56578-56881 | XP_005175924.1 | kazal domain | 78 | 59 | 6608.54 | −3.0 | 4.98 |
| MdMuslin6 | AQPM01019378.1:57384-57671 | XP_005175923.1 | kazal domain | 73 | 53 | 5905.82 | 5.0 | 8.73 |
| MdMuslin7 | NDYK01036986.1:1550-1765 | kazal domain | 70 | 51 | 5592.32 | −1.3 | 5.12 | |
| MdMuslin8 | NDYK01185191.1:567-854 | XP_011294170.1 | kazal domain | 75 | 56 | 6360.27 | 4.2 | 8.52 |
| MdMuslin9 | XP_005175922.1 | kazal domain | 75 | 56 | 6165.88 | −1.3 | 5.27 | |
| MdMuslin10 | AQPM01042957.1:886-1101 | XP_005175386.1 | kazal domain | 72 | 48 | 5340.00 | 0.7 | 7.61 |
| MdMuslin11 | NDYK01172454.1:1382-1630 | XP_005190386.1 | kazal domain | 83 | 63 | 6751.57 | −1.0 | 5.97 |
| MdMuslin12 | NDYK01221433.1:787-1002 | XP_005188808.1 | kazal domain | 82 | 49 | 5613.51 | 3.7 | 8.50 |
| MdMuslin13 | XP_005178654.1 | kazal domain | 136 | 117 | 12354.50 | 4.0 | 8.55 | |
| MdMuslin14 | XP_005188224.1 | kazal domain | 79 | 59 | 6456.06 | −5.3 | 4.49 | |
| MdMuslin15-1 | XP_005188061.1 | kazal domain | 154 | 76 | 8566.70 | −3.3 | 4.77 | |
| MdMuslin15-2 | kazal domain | 57 | 6219.32 | 1.7 | 7.98 | |||
| MdMuslin16-1 | AQPM01094907.1:8864-9376 | kazal domain | 148 | 77 | 8667.80 | −3.3 | 4.77 | |
| MdMuslin16-2 | kazal domain | 50 | 5537.53 | 1.7 | 7.98 | |||
| MdMuslin17 | NDYK01122947.1:2962-3420 | XP_005190966.1 | kazal domain | 88 | 67 | 7092.01 | 0.7 | 7.61 |
| MdMuslin18 | XP_005190967.1 | kazal domain | 98 | 70 | 7304.27 | 4.7 | 8.80 | |
| MdMuslin19 | NDYK01014763:1939-2223 | kazal domain | 95 | 67 | 7112.23 | 6.7 | 9.30 | |
| MdMuslin20 | XP_019893634.1 | kazal domain | 83 | 62 | 6943.64 | −8.0 | 4.16 | |
| MdMuslin21 | XP_011296278.1 | kazal domain | 98 | 77 | 8811.17 | 4.2 | 8.52 | |
| MdMuslin22 | XP_005188497.1 | kazal domain | 91 | 72 | 7752.70 | 2.0 | 7.98 | |
| MdMuslin23-1 | XP_005190965.2 | kazal domain | 136 | 57 | 6848.86 | 3.0 | 8.27 | |
| MdMuslin23-2 | kazal domain | 52 | 5968.77 | 0.7 | 7.61 | |||
| MdMuslin24-1 | XP_019894976.1 | kazal domain | 135 | 61 | 7210.20 | −1.3 | 5.41 | |
| MdMuslin24-2 | kazal domain | 52 | 6100.76 | −0.5 | 6.72 | |||
| MdMuslin25 | AQPM01000599.1:6069-6748 | kazal domain | 87 | 64 | 7410.43 | 10.4 | 10.09 | |
| MdMuslin26 | NDYK01190177.1:7-310 | kazal domain | 77 | 56 | 6388.29 | 5.2 | 8.65 | |
| MdMuslin27 | AQPM01000601.1:7182-7517 | XP_005190993.1 | kazal domain | 87 | 64 | 7268.36 | 9.7 | 10.89 |
| MdMuslin28 | AQPM01000604.1:11632-11959 | XP_011295627.1 | kazal domain | 89 | 66 | 7438.50 | 7.9 | 9.30 |
| MdMuslin29 | AQPM01000605.1:5737-6069 | kazal domain | 87 | 64 | 7267.41 | 8.9 | 9.30 | |
| MdMuslin30 | XP_005190994.1 | kazal domain | 90 | 67 | 7619.84 | 9.9 | 9.51 | |
| MdMuslin31 | NDYK01044765.1:1300-1610 | kazal domain | 83 | 61 | 7115.23 | 10.9 | 10.61 | |
| MdMuslin32 | AQPM01000603.1:4008-4322 | kazal domain | 84 | 59 | 6658.82 | 5.9 | 8.66 | |
| MdMuslin33 | AQPM01000603.1:639-963 | kazal domain | 89 | 66 | 7007.07 | 7.1 | 8.90 | |
| MdSVWC1 | XP_005183514.1 | svwc | 149 | 130 | 14382.45 | −4.3 | 5.08 | |
| MdSVWC2 | XP_011295325.1 | svwc | 102 | 81 | 8733.00 | −1.1 | 5.88 | |
| MdSVWC3 | NDYK01201822.1:136-580 | XP_005175282.1 | svwc | 102 | 81 | 8741.00 | −1.1 | 5.88 |
| MdSVWC4 | XP_005189656.1 | svwc | 134 | 103 | 11455.80 | −3.3 | 5.01 | |
| MdSVWC5 | AQPM01015706.1:698-1200 | svwc | 112 | 94 | 10684.30 | −1.6 | 6.25 | |
| MdSVWC6 | XP_005175408.1 | svwc | 120 | 100 | 11347.19 | 0.4 | 7.19 | |
| MdSVWC7 | AQPM01092559.1:3374-5544 | XP_005187657.1 | svwc | 182 | 163 | 19163.26 | 1.9 | 7.87 |
| MdSVWC8 | AQPM01024958.1:41-530 | XP_005190002.1 | svwc | 113 | 94 | 10879.12 | −2.6 | 5.78 |
| MdSVWC9 | AQPM01015051.1: 3997-5502 | XP_005175283.1 | svwc | 111 | 87 | 10386.73 | −2.1 | 6.34 |
| MdSVWC10 | AQPM01015049.1:461-992 | svwc | 100 | 80 | 9437.82 | 7.4 | 8.88 | |
| MdSVWC11 | XP_011295271.1 | svwc | 104 | 84 | 9971.32 | 0.6 | 7.26 | |
| MdSVWC12 | AQPM01015050.1:115-2228 | XP_005175281.1 | svwc | 104 | 79 | 9215.33 | −1.6 | 6.25 |
| MdSVWC13 | XP_005175284.1 | svwc | 88 | 68 | 7510.65 | 3.9 | 8.29 | |
| MdSVWC14 | AQPM01081312.1:14881-15505 | svwc | 102 | 79 | 8904.37 | 0.9 | 7.52 | |
| MdSVWC15 | NW_004765359.1:113654-114278 | JZ121963.1 | svwc | 95 | 72 | 8095.36 | −0.1 | 6.91 |
| MdSVWC16 | XP_005184317.1 | svwc | 124 | 96 | 10843.56 | 0.2 | 7.09 | |
| MdSVWC17 | AQPM01092395.1:1108-1569 | XP_005187600.1 | svwc | 121 | 102 | 11254.72 | −0.4 | 6.86 |
| MdSVWC18 | XP_005180761.1 | svwc | 107 | 88 | 9682.02 | 4.2 | 8.31 | |
| MdSVWC19 | AQPM01056437.1:2396-2894 | XP_011290794.1 | svwc | 113 | 89 | 9482.15 | −0.8 | 6.44 |
| MdSVWC20 | AQPM01095428.1:3680-4100 | XP_005188179.1 | svwc | 102 | 84 | 9247.47 | −5.1 | 4.75 |
| MdSVWC21 | AQPM01060615.1:2747-3172 | XP_005180301.1 | svwc | 103 | 85 | 9126.49 | −1.6 | 6.25 |
| MdSVWC22 | AQPM01095425.1:2644-3102 | XP_005188177.1 | svwc | 113 | 95 | 10296.72 | −4.6 | 5.10 |
| MdSVWC23 | XP_011294423.1 | svwc | 120 | 102 | 11126.55 | −4.6 | 5.10 | |
| MdSVWC24 | AQPM01095428.1:13247-13749 | XP_011294424.1 | svwc | 105 | 85 | 9320.74 | 4.2 | 8.31 |
| MdSVWC25 | AQPM01095423.1:787-1232 | XP_005188176.1 | svwc | 106 | 84 | 8913.27 | 3.2 | 8.12 |
| MdSVWC26 | AQPM01019310.1:3498-3948 | XP_005175918.1 | svwc | 106 | 84 | 9408.71 | −0.6 | 6.72 |
| MdSVWC27 | AQPM01095427.1:815-1458 | XP_005188181.1 | svwc | 115 | 95 | 10876.26 | −6.9 | 4.77 |
| MdSVWC28 | AQPM01095425.1:13938-14587 | svwc | 115 | 95 | 10891.33 | −7.8 | 4.67 | |
| MdSVWC29-1 | XP_005188180.2 | svwc | 137 | 117 | 13288.15 | −7.9 | 4.60 | |
| MdSVWC29-2 | svwc | 101 | 83 | 9053.29 | −0.8 | 6.48 | ||
| MdCrustin1 | AQPM01030484.1:548-1269 | wappin domain | 100 | 67 | 7493.40 | 2.9 | 8.10 | |
| MdCrustin2 | XP_011295532.1 | wappin domain | 92 | 67 | 7493.40 | 2.9 | 8.10 | |
| MdCrustin3 | XP_005190815.1 | wappin domain | 115 | 94 | 9819.93 | 5.8 | 8.37 | |
| MdCrustin4 | XP_011295531.1 | wappin domain | 120 | 95 | 10541.92 | 6.8 | 8.48 | |
| MdCecropin1 # | ABB17292.1 | α-helix | 63 | 40 | 4271.97 | 5.1 | 10.56 | |
| MdCecropin2 # | AQPM01058001.1:775-1030 | XP_005179713.1 | α-helix | 64 | 41 | 4342.06 | 6.1 | 10.66 |
| MdCecropin3 # | AQPM01058001.1:8963-9222 | XP_005179700.1 | α-helix | 64 | 41 | 4386.11 | 6.1 | 10.66 |
| MdCecropin4 # | AQPM01058004.1:2369-2661 | XP_019890986.1 | α-helix | 63 | 40 | 4257.94 | 5.1 | 10.56 |
| MdCecropin5 | NDYK01010340.1:75-311 | α-helix | 64 | 41 | 4461.10 | 4.2 | 10.94 | |
| MdCecropin6 # | AQPM01058004.1:5352-5771 | XP_005179717.1 | α-helix | 64 | 41 | 4370.11 | 6.1 | 10.66 |
| MdCecropin7 # | AQPM01058000.1:20546-20804 | XP_005179712.1 | α-helix | 63 | 41 | 4356.05 | 6.1 | 11.12 |
| MdCecropin8 # | AQPM01058001.1:4386-4681 | AXG50148.1 | α-helix | 64 | 41 | 4356.09 | 6.1 | 10.66 |
| MdCecropin9 # | AQPM01058006.1:1083-1338 | XP_005179718.1 | α-helix | 64 | 41 | 4461.10 | 4.2 | 10.94 |
| MdCecropin10 # | AQPM01058008.1:3578-4516 | XP_005179719.1 | α-helix | 62 | 41 | 4464.06 | 3.2 | 10.28 |
| MdCecropin11 # | AQPM01058004.1:928-1210 | AIW52264.1 | α-helix | 64 | 41 | 4356.09 | 6.1 | 10.66 |
| MdCecropin12 | JZ121081.1 | α-helix | 63 | 40 | 4227.91 | 5.1 | 10.56 | |
| MdCecropin13 | ES608288.1 | α-helix | 64 | 41 | 4341.12 | 6.1 | 10.66 | |
| MdCecropin14 # | AQPM01100428.1:3910-4122 | XP_011294761.1 | α-helix | 69 | 44 | 4546.24 | 2.9 | 9.34 |
| MdCecropin15 | AQPM01100427.1:1792-2047 | XP_019894290.1 | α-helix | 69 | 44 | 4518.23 | 3.9 | 9.72 |
| MdCecropin16 | AQPM01100428.1:8445-8711 | α-helix | 69 | 44 | 4560.27 | 2.9 | 9.34 | |
| MdDiptericin1 # | FJ748596.1:148-344 | ACN61637.1 | Proline and Glycine rich | 99 | 79 | 8721.39 | 1.4 | 8.50 |
| MdDiptericin2 # | FJ794602.1:25-321 | ACO35257.1 | Proline and Glycine rich | 99 | 79 | 8721.39 | 1.4 | 8.50 |
| MdDiptericin3 # | KM205631.151-347 | Proline and Glycine rich | 99 | 79 | 8596.58 | 3.5 | 8.69 | |
| MdDiptericin4 # | FJ795370.1:65-364 | ACN93798.1 | Proline and Glycine rich | 99 | 79 | 8725.34 | 1.4 | 8.50 |
| MdDiptericinD # | AQPM01092243.1:221-1086 | NP_001295957.2 | Proline and Glycine rich | 99 | 79 | 8770.47 | 1.4 | 8.41 |
| MdDiptericinD1 # | AQPM01092241.1:4245-4608 | XP_005187575.1 | Proline and Glycine rich | 99 | 79 | 8711.36 | 1.4 | 8.50 |
| MdDomesticin1 # | AHA56721.1 | Proline rich | 65 | 40 | 4583.33 | 6.9 | 11.41 | |
| MdDomesticin2 | AQPM01056449.1:10440-12589 | Proline rich | 65 | 40 | 4525.20 | 5.9 | 10.98 | |
| MdEdin1 | AQPM01067938.1:2033-2341 | Glycine rich | 102 | 62 | 6987.33 | −0.9 | 6.67 | |
| MdEdin2 | AQPM01067938.1:5150-5500 | Glycine rich | 116 | 65 | 7321.74 | 2.4 | 9.20 | |
| MdEdin3 | AQPM01067936.1:1742-2049 | Glycine rich | 101 | 61 | 6827.11 | −1.9 | 6.34 | |
| MdEdin4 | AQPM01067936.1:4517-4852 | Glycine rich | 120 | 65 | 7331.79 | 3.6 | 9.70 | |
| MdEdin5 | JZ121894.1 | Glycine rich | 116 | 65 | 7359.79 | 2.4 | 9.25 | |
| MdEdin6 | AQPM01067939.1:3916-4450 | Glycine rich | 177 | 127 | 13760.56 | 1.1 | 7.66 | |
| MdEdin7 | NDYK01101123.1:1387-1910 | Glycine rich | 174 | 127 | 13721.52 | 1.9 | 8.50 | |
| MdEdin8 | AQPM01067938.1:9198-9731 | Glycine rich | 177 | 127 | 13752.54 | 0.7 | 7.47 | |
| MdEdin9 | AQPM01067938.1:15033-15560 | Glycine rich | 174 | 125 | 13612.44 | 3.6 | 9.34 | |
| MdEdin10 # | AFP64086.1 | Glycine rich | 175 | 127 | 13740.57 | 1.7 | 8.50 | |
| MdAttacinA1 # | XP_011296530.1 | Glycine rich | 208 | 188 | 20002.01 | 6.9 | 9.66 | |
| MdAttacinA2 | AQPM01013309.1:3192-3887 | XP_019890218.1 | Glycine rich | 208 | 188 | 19613.34 | 6.9 | 9.81 |
| MdAttacinA3 # | AAY59540.1 | Glycine rich | 208 | 188 | 19688.41 | 6.9 | 9.81 | |
| MdAttacinA4 # | AAR23786.1 | Glycine rich | 208 | 188 | 19672.41 | 6.9 | 9.81 | |
| MdAttacinA5 | XP_019890219.1 | Glycine rich | 208 | 188 | 19718.44 | 6.9 | 9.81 | |
| MdAttacinC1 # | AQPM01059487.1:2038-2826 | Proline and Glycine rich | 241 | 192 | 20420.06 | 2.9 | 8.92 | |
| MdAttacinC2 # | ACO35258.1 | Proline and Glycine rich | 241 | 192 | 20334.91 | 1.9 | 8.41 | |
| MdAttacinC3-1 | XP_005180079.2 | Proline and Glycine rich | 250 | 201 | 21385.12 | 5.1 | 9.34 | |
| MdAttacinC3-2 | Proline and Glycine rich | 241 | 192 | 20449.11 | 4.1 | 9.23 | ||
| MdAttacinC4 # | AQPM01059487.1:7828-8614 | XP_005180076.1 | Proline and Glycine rich | 241 | 192 | 20383.93 | 3.1 | 8.92 |
| MdAttacinC5 # | NDYK01054543.1:3986-4696 | Proline and Glycine rich | 241 | 192 | 19973.48 | 5.1 | 9.34 | |
| MdAttacinC6 | AQPM01059487.1:3997-5074 | Proline and Glycine rich | 241 | 192 | 20349.91 | 3.1 | 8.92 | |
| MdAttacinC7 | JZ121354.1:31-753 | Proline and Glycine rich | 241 | 192 | 20384.91 | 1.7 | 8.41 | |
| MdAttacinD1 | XP_011296538.1 | Glycine rich | 181 | 181 | 19122.94 | 8.1 | 9.91 | |
| MdAttacinD2 | XP_005178516.1 | Glycine rich | 189 | 189 | 19412.19 | 5.3 | 9.41 | |
| MdAttacinD3 # | NDYK01109436.1:1208-1899 | XP_005178550.1 | Glycine rich | 191 | 191 | 19712.79 | 7.8 | 9.70 |
| MdAttacinD4 # | AFP64340.1 | Glycine rich | 197 | 197 | 21110.83 | 6.1 | 9.65 | |
References
- Hoffmann, J.A. Innate immunity of insects. Curr. Opin. Immunol. 1995, 7, 4–10. [Google Scholar] [CrossRef]
- Stork, N.E.; McBroom, J.; Gely, C.; Hamilton, A.J. New approaches narrow global species estimates for beetles, insects, and terrestrial arthropods. Proc. Natl. Acad. Sci. USA 2015, 112, 7519–7523. [Google Scholar] [CrossRef] [Green Version]
- Lemaitre, B.; Hoffmann, J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007, 25, 697–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flores-Villegas, A.L.; Salazar-Schettino, P.M.; Cordoba-Aguilar, A.; Gutierrez-Cabrera, A.E.; Rojas-Wastavino, G.E.; Bucio-Torres, M.I.; Cabrera-Bravo, M. Immune defence mechanisms of triatomines against bacteria, viruses, fungi and parasites. Bull. Entomol. Res. 2015, 105, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Hahm, K.S. Antimicrobial peptides (AMPs): Peptide structure and mode of action. J. Biochem. Mol. Biol. 2005, 38, 507–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imler, J.L.; Bulet, P. Antimicrobial peptides in Drosophila: Structures, activities and gene regulation. Chem. Immunol. Allergy 2005, 86, 1–21. [Google Scholar] [PubMed]
- Zhang, Z.T.; Zhu, S.Y. Drosomycin, an essential component of antifungal defence in Drosophila. Insect. Mol. Biol. 2009, 18, 549–556. [Google Scholar] [CrossRef]
- Hanson, M.A.; Lemaitre, B. New insights on Drosophila antimicrobial peptide function in host defense and beyond. Curr. Opin. Immunol. 2020, 62, 22–30. [Google Scholar] [CrossRef]
- Bulet, P.; Hetru, C.; Dimarcq, J.L.; Hoffmann, D. Antimicrobial peptides in insects; structure and function. Dev. Comp. Immunol. 1999, 23, 329–344. [Google Scholar] [CrossRef]
- Kaushal, A.; Gupta, K.; Shah, R.; van Hoek, M.L. Antimicrobial activity of mosquito cecropin peptides against Francisella. Dev Comp. Immunol. 2016, 63, 171–180. [Google Scholar] [CrossRef]
- Casteels, P.; Ampe, C.; Jacobs, F.; Vaeck, M.; Tempst, P. Apidaecins: Antibacterial peptides from honeybees. EMBO J. 1989, 8, 2387–2391. [Google Scholar] [CrossRef]
- Charlet, M.; Lagueux, M.; Reichhart, J.M.; Hoffmann, D.; Braun, A.; Meister, M. Cloning of the gene encoding the antibacterial peptide drosocin involved in Drosophila immunity. Eur. J. Biochem. 1996, 241, 699–706. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Taniai, K.; Hara, S.; Kadonookuda, K.; Kato, Y.; Yamamoto, M.; Xu, J.; Choi, S.K.; Debnath, N.C.; Choi, H.K.; et al. cDNA cloning and gene expression of lebocin, a novel member of antibacterial peptides from the silkworm, Bombyx mori. Biochem. Biophys. Res. Commun. 1995, 214, 271–278. [Google Scholar] [CrossRef]
- Gao, B.; Zhu, S. The drosomycin multigene family: Three-disulfide variants from Drosophila takahashii possess antibacterial activity. Sci. Rep. 2016, 6, 32175–32186. [Google Scholar] [CrossRef] [Green Version]
- Thevissen, K.; Kristensen, H.H.; Thomma, B.P.; Cammue, B.P.; François, I.E. Therapeutic potential of antifungal plant and insect defensins. Drug Discov. Today 2007, 12, 966–971. [Google Scholar] [CrossRef] [PubMed]
- Mylonakis, E.; Podsiadlowski, L.; Muhammed, M.; Vilcinskas, A. Diversity, evolution and medical applications of insect antimicrobial peptides. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016, 371, 1695–1705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christophides, G.K.; Zdobnov, E.; Barillas-Mury, C.; Birney, E.; Blandin, S.; Blass, C.; Brey, P.T.; Collins, F.H.; Danielli, A.; Dimopoulos, G.; et al. Immunity-related genes and gene families in Anopheles gambiae. Science 2002, 298, 159–165. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, H.; Ishibashi, J.; Fujita, K.; Nakajima, Y.; Sagisaka, A.; Tomimoto, K.; Suzuki, N.; Yoshiyama, M.; Kaneko, Y.; Iwasaki, T.; et al. A genome-wide analysis of genes and gene families involved in innate immunity of Bombyx mori. Insect. Biochem. Mol. Biol. 2008, 38, 1087–1110. [Google Scholar] [CrossRef]
- Waterhouse, R.M.; Kriventseva, E.V.; Meister, S.; Xi, Z.; Alvarez, K.S.; Bartholomay, L.C.; Barillas-Mury, C.; Bian, G.; Blandin, S.; Christensen, B.M.; et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science 2007, 316, 1738–1743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, C.; Gao, B.; Fang, Q.; Ye, G.; Zhu, S. Antimicrobial peptide-like genes in Nasonia vitripennis: A genomic perspective. BMC Genomics 2010, 11, 187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, Y.; Zheng, D.; Yao, B.; Cai, Z.; Zhao, Z.; Wu, S.; Cong, P.; Yang, D. A novel bioconversion for value-added products from food waste using Musca domestica. Waste Manag. 2017, 61, 455–460. [Google Scholar] [CrossRef]
- Scott, J.G.; Warren, W.C.; Beukeboom, L.W.; Bopp, D.; Clark, A.G.; Giers, S.D.; Hediger, M.; Jones, A.K.; Kasai, S.; Leichter, C.A.; et al. Genome of the house fly, Musca domestica L. a global vector of diseases with adaptations to a septic environment. Genome Biol. 2014, 15, 466–482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, S.; Gao, B.; Tytgat, J. Phylogenetic distribution, functional epitopes and evolution of the CSαβ superfamily. Cell Mol. Life Sci. 2005, 62, 2257–2269. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, S.; Gao, B.; Peigneur, S.; Tytgat, J. How a scorpion toxin selectively captures a prey sodium channel: The molecular and evolutionary basis uncovered. Mol. Biol. Evol. 2020, 37, 3149–3164. [Google Scholar] [CrossRef]
- Pierce, B.G.; Wiehe, K.; Hwang, H.; Kim, B.H.; Vreven, T.; Weng, Z. ZDOCK server: Interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics 2014, 30, 1771–1773. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Kaminski, G.A.; Friesner, R.A.; Tirado-Rives, J.; Jorgensen, W.L. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J. Phys. Chem. B 2001, 105, 6474–6487. [Google Scholar] [CrossRef]
- Zhu, S.; Gao, B. Positive selection in cathelicidin host defense peptides: Adaptation to exogenous pathogens or endogenous receptors? Heredity 2017, 118, 453–465. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Nielsen, R.; Goldman, N.; Pedersen, A.M.K. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 2000, 155, 431–449. [Google Scholar]
- Yang, Z.; Wong, W.S.W.; Nielsen, R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005, 22, 1107–1118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colell, E.A.; Iserte, J.A.; Simonetti, F.L.; Marino-Buslje, C. MISTIC2: Comprehensive server to study coevolution in protein families. Nucleic Acids Res. 2018, 46, W323–W328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, W.; Gao, B.; Zhu, S. Did cis- and trans-defensins derive from a common ancestor? Immunogenetics 2019, 71, 61–69. [Google Scholar] [CrossRef]
- Cociancich, S.; Ghazi, A.; Hetru, C.; Hoffmann, J.A.; Letellier, L. Insect defensin, an inducible antibacterial peptide, forms voltage-dependent channels in Micrococcus luteus. J. Biol. Chem. 1993, 268, 19239–19245. [Google Scholar] [CrossRef]
- Lee, Y.S.; Yun, E.K.; Jang, W.S.; Kim, I.; Lee, J.H.; Park, S.Y.; Ryu, K.S.; Seo, S.J.; Kim, C.H.; Lee, I.H. Purification, cDNA cloning and expression of an insect defensin from the great wax moth, Galleria mellonella. Insect. Mol. Biol. 2004, 13, 65–72. [Google Scholar] [CrossRef]
- Andoh, M.; Ueno, T.; Kawasaki, K. Tissue-dependent induction of antimicrobial peptide genes after body wall injury in house fly (Musca domestica) larvae. Drug Discov. Ther. 2018, 12, 355–362. [Google Scholar] [CrossRef] [Green Version]
- Yount, N.Y.; Yeaman, M.R. Multidimensional signatures in antimicrobial peptides. Proc. Natl. Acad. Sci. USA 2004, 101, 7363–7368. [Google Scholar] [CrossRef] [Green Version]
- Koehbach, J. Structure-Activity Relationships of Insect. Defensins. Front. Chem. 2007, 5, 45–54. [Google Scholar] [CrossRef] [Green Version]
- Zhu, S. Discovery of six families of fungal defensin-like peptides provides insights into origin and evolution of the CSαβ defensins. Mol. Immunol. 2008, 45, 828–838. [Google Scholar] [CrossRef]
- Laskowski, M., Jr.; Kato, I. Protein inhibitors of proteinases. Annu. Rev. Biochem. 1980, 49, 593–626. [Google Scholar] [CrossRef] [PubMed]
- Kanost, M.R. Serine proteinase inhibitors in arthropod immunity. Dev. Comp. Immunol. 1999, 23, 291–301. [Google Scholar] [CrossRef]
- McCrudden, M.T.; Dafforn, T.R.; Houston, D.F.; Turkington, P.T.; Timson, D.J. Functional domains of the human epididymal protease inhibitor, eppin. FEBS J. 2008, 275, 1742–1750. [Google Scholar] [CrossRef]
- Fröbius, A.C.; Kanost, M.R.; Götz, P.; Vilcinskas, A. Isolation and characterization of novel inducible serine protease inhibitors from larval hemolymph of the greater wax moth Galleria mellonella. Eur. J. Biochem. 2000, 267, 2046–2053. [Google Scholar] [CrossRef] [PubMed]
- Nirmala, X.; Kodrík, D.; Žurovec, M.; Sehnal, F. Insect silk contains both a Kunitz-type and a unique Kazal-typeproteinase inhibitor. Eur. J. Biochem. 2001, 268, 2064–2073. [Google Scholar] [CrossRef] [Green Version]
- de Magalhaes, M.T.Q.; Mambelli, F.S.; Santos, B.P.O.; Morais, S.B.; Oliveira, S.C. Serine protease inhibitors containing a Kunitz domain: Their role in modulation of host inflammatory responses and parasite survival. Microbes Infect. 2018, 20, 606–609. [Google Scholar] [CrossRef]
- Watanabe, R.M.; Soares, T.S.; Morais-Zani, K.; Tanaka-Azevedo, A.M.; Maciel, C.; Capurro, M.L.; Torquato, R.J.; Tanaka, A.S. A novel trypsin Kazal-type inhibitor from Aedes aegypti with thrombin coagulant inhibitory activity. Biochimie 2010, 92, 933–939. [Google Scholar] [CrossRef]
- Niimi, T.; Yokoyama, H.; Goto, A.; Beck, K.; Kitagawa, Y. A Drosophila gene encoding multiple splice variants of Kazal-type serine protease inhibitor-like proteins with potential destinations of mitochondria, cytosol and the secretory pathway. Eur. J. Biochem. 1999, 266, 282–292. [Google Scholar] [CrossRef] [Green Version]
- Brillard-Bourdet, M.; Hamdaoui, A.; Hajjar, E.; Boudier, C.; Reuter, N.; Ehret-Sabatier, L.; Bieth, J.G.; Gauthier, F. A novel locust (Schistocerca gregaria) serine protease inhibitor with a high affinity for neutrophil elastase. Biochem. J. 2006, 400, 467–476. [Google Scholar] [CrossRef] [Green Version]
- Kumaresan, V.; Harikrishnan, R.; Arockiaraj, J. A potential Kazal-type serine protease inhibitor involves in kinetics of protease inhibition and bacteriostatic activity. Fish Shellfish Immunol. 2015, 42, 430–438. [Google Scholar] [CrossRef]
- Gebhard, L.G.; Carrizo, F.U.; Stern, A.L.; Burgardt, N.I.; Faivovich, J.; Lavilla, E.; Ermacora, M.R. A Kazal prolyl endopeptidase inhibitor isolated from the skin of Phyllomedusa sauvagii. Eur. J. Biochem. 2004, 271, 2117–2126. [Google Scholar] [CrossRef] [Green Version]
- Han, F.; Lu, A.; Yuan, Y.; Huang, W.; Beerntsen, B.T.; Huang, J.; Ling, E. Characterization of an entomopathogenic fungi target integument protein, Bombyx mori single domain von Willebrand factor type C, in the silkworm, Bombyx mori. Insect. Mol. Biol. 2017, 26, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Smit, A.B.; de Jong-Brink, M.; Li, K.W.; Sassen, M.M.J.; Spijker, S.; van Elk, R.; Buijs, S.P.; van Minnen, J.; van Kesteren, R.E. Granularin, a novel molluscan opsonin comprising a single vWF type C domain is up-regulated during parasitation. FEBS J. 2004, 18, 845–847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, V.J.; Fernandes, J.M.; Kemp, G.D.; Hauton, C. Crustins: Enigmatic WAP domain-containing antibacterial proteins from crustaceans. Dev. Comp. Immunol. 2008, 32, 758–772. [Google Scholar] [CrossRef] [Green Version]
- Vargas-Albores, F.; Martinez-Porchas, M. Crustins are distinctive members of the WAP-containing protein superfamily: An improved classification approach. Dev. Comp. Immunol. 2017, 76, 9–17. [Google Scholar] [CrossRef]
- Afsal, V.V.; Antony, S.P.; Sathyan, N.; Philip, R. Molecular characterization and phylogenetic analysis of two antimicrobial peptides: Anti-lipopolysaccharide factor and crustin from the brown mud crab, Scylla serrata. Results Immunol. 2011, 1, 6–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brockton, V.; Hammond, J.A.; Smith, V.J. Gene characterisation, isoforms and recombinant expression of carcinin, an antibacterial protein from the shore crab, Carcinus maenas. Mol. Immunol. 2007, 44, 943–949. [Google Scholar] [CrossRef] [Green Version]
- Antony, S.P.; Singh, I.S.; Sudheer, N.S.; Vrinda, S.; Priyaja, P.; Philip, R. Molecular characterization of a crustin-like antimicrobial peptide in the giant tiger shrimp, Penaeus monodon, and its expression profile in response to various immunostimulants and challenge with WSSV. Immunobiology 2011, 216, 184–194. [Google Scholar] [CrossRef]
- Yang, L.; Niu, S.; Gao, J.; Zuo, H.; Yuan, J.; Weng, S.; He, J.; Xu, X. A single WAP domain (SWD)-containing protein with antiviral activity from Pacific white shrimp Litopenaeus vannamei. Fish Shellfish Immunol. 2018, 73, 167–174. [Google Scholar] [CrossRef]
- Zhang, J.; Li, F.; Wang, Z.; Xiang, J. Cloning and recombinant expression of a crustin-like gene from Chinese shrimp, Fenneropenaeus chinensis. J. Biotechnol. 2007, 127, 605–614. [Google Scholar] [CrossRef]
- Hagiwara, K.; Kikuchi, T.; Endo, Y.; Usui, K.; Takahashi, M.; Shibata, N.; Kusakabe, T.; Xin, H.; Hoshi, S.; Miki, M.; et al. Mouse SWAM1 and SWAM2 are antibacterial proteins composed of a single whey acidic protein motif. J. Immunol. 2003, 170, 1973–1979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nair, D.G.; Fry, B.G.; Alewood, P.; Kumar, P.P.; Kini, R.M. Antimicrobial activity of omwaprin, a new member of the waprin family of snake venom proteins. Biochem. J. 2007, 402, 93–104. [Google Scholar] [CrossRef]
- Steiner, H.; Hultmark, D.; Engström, A.; Bennich, H.; Boman, H.G. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 1981, 182, 246–248. [Google Scholar] [CrossRef]
- Peng, J.; Wu, Z.; Liu, W.; Long, H.; Zhu, G.; Guo, G.; Wu, J. Antimicrobial functional divergence of the cecropin antibacterial peptide gene family in Musca domestica. Parasit. Vectors 2019, 12, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Boulanger, N.; Munks, R.J.; Hamilton, J.V.; Vovelle, F.; Brun, R.; Lehane, M.J.; Bulet, P. Epithelial innate immunity. A novel antimicrobial peptide with antiparasitic activity in the blood-sucking insect Stomoxys calcitrans. J. Biol. Chem. 2002, 277, 49921–49926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vizioli, J.; Bulet, P.; Charlet, M.; Lowenberger, C.; Blass, C.; Muller, H.M.; Dimopoulos, G.; Hoffmann, J.; Kafatos, F.C.; Richman, A. Cloning and analysis of a cecropin gene from the malaria vector mosquito, Anopheles gambiae. Insect. Mol. Biol. 2000, 9, 75–84. [Google Scholar] [CrossRef] [Green Version]
- Ekengren, S.; Hultmark, D. Drosophila cecropin as an antifungal agent. Insect. Biochem. Mol. Biol. 1999, 29, 965–972. [Google Scholar] [CrossRef]
- Okada, M.; Natori, S. Primary structure of sarcotoxin I, an antibacterial protein induced in the hemolymph of Sarcophaga peregrina (flesh fly) larvae. J. Biol. Chem. 1985, 260, 7174–7177. [Google Scholar] [CrossRef]
- Ouyang, L.; Xu, X.; Freed, S.; Gao, Y.; Yu, J.; Wang, S.; Ju, W.; Zhang, Y.; Jin, F. Cecropins from Plutella xylostella and their interaction with Metarhizium anisopliae. PLoS ONE 2015, 10, e0142451. [Google Scholar] [CrossRef] [PubMed]
- Saito, A.; Ueda, K.; Imamura, M.; Atsumi, S.; Tabunoki, H.; Miura, N.; Watanabe, A.; Kitami, M.; Sato, R. Purification and cDNA cloning of a cecropin from the longicorn beetle, Acalolepta luxuriosa. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2005, 142, 317–323. [Google Scholar] [CrossRef]
- Kim, J.K.; Lee, E.; Shin, S.; Jeong, K.W.; Lee, J.Y.; Bae, S.Y.; Kim, S.H.; Lee, J.; Kim, S.R.; Lee, D.G.; et al. Structure and function of papiliocin with antimicrobial and anti-inflammatory activities isolated from the swallowtail butterfly, Papilio xuthus. J. Biol. Chem. 2011, 286, 41296–41311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, E.; Jeong, K.W.; Lee, J.; Shin, A.; Kim, J.-K.; Lee, J.; Lee, D.G.; Kim, Y. Structure-activity relationships of cecropin-like peptides and their interactions with phospholipid membrane. BMB Rep. 2013, 46, 282–287. [Google Scholar] [CrossRef]
- Yagi-Utsumi, M.; Yamaguchi, Y.; Boonsri, P.; Iguchi, T.; Okemoto, K.; Natori, S.; Kato, K. Stable isotope-assisted NMR characterization of interaction between lipid A and sarcotoxin IA, a cecropin-type antibacterial peptide. Biochem. Biophys. Res. Commun. 2013, 431, 136–140. [Google Scholar] [CrossRef] [PubMed]
- Okemoto, K.; Nakajima, Y.; Fujioka, T.; Natori, S. Participation of two N-terminal residues in LPS neutralizing activity of sarcotoxin IA. J. Biochem. 2002, 131, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.; Shin, S.Y.; Lee, S.; Kang, J.H.; Kim, S.D.; Ryu, P.D.; Hahm, K.S.; Kim, Y. Role of the hinge region and the tryptophan residue in the synthetic antimicrobial peptides, cecropin A(1–8)-magainin 2(1–12) and its analogues, on their antibiotic activities and structures. Biochemistry 2000, 39, 11855–11864. [Google Scholar] [CrossRef] [PubMed]
- Pei, Z.; Sun, X.; Tang, Y.; Wang, K.; Gao, Y.; Ma, H. Cloning, expression, and purification of a new antimicrobial peptide gene from Musca domestica larva. Gene 2014, 549, 41–45. [Google Scholar] [CrossRef]
- Tang, T.; Li, X.; Yang, X.; Yu, X.; Wang, J.; Liu, F.; Huang, D. Transcriptional response of Musca domestica larvae to bacterial infection. PLoS ONE 2014, 9, e104867. [Google Scholar] [CrossRef] [Green Version]
- Scocchi, M.; Tossi, A.; Gennaro, R. Proline-rich antimicrobial peptides: Converging to a non-lytic mechanism of action. Cell Mol. Life Sci. 2011, 68, 2317–2330. [Google Scholar] [CrossRef] [PubMed]
- Dimarcq, J.L.; Zachary, D.; Hoffmann, J.A.; Hoffmann, D.; Reichhart, J.M. Insect immunity: Expression of the two major inducible antibacterial peptides, defensin and diptericin, in Phormia terranovae. EMBO J. 1990, 9, 2507–2515. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Cho, K.S.; Lee, J.; Yoo, J.; Lee, J.; Chung, J. Diptericin-like protein: An immune response gene regulated by the anti-bacterial gene induction pathway in Drosophila. Gene 2001, 271, 233–238. [Google Scholar] [CrossRef]
- Kim, C.H.; Muturi, E.J. Effect of larval density and Sindbis virus infection on immune responses in Aedes aegypti. J. Insect. Physiol. 2013, 59, 604–610. [Google Scholar] [CrossRef]
- Vanha-Aho, L.M.; Anderl, I.; Vesala, L.; Hultmark, D.; Valanne, S.; Ramet, M. Edin expression in the fat body is required in the defense against parasitic wasps in Drosophila melanogaster. PLoS Pathog. 2015, 11, e1004895. [Google Scholar] [CrossRef] [PubMed]
- Verleyen, P.; Baggerman, G.; D’Hertog, W.; Vierstraete, E.; Husson, S.J.; Schoofs, L. Identification of new immune induced molecules in the haemolymph of Drosophila melanogaster by 2D-nanoLC MS/MS. J. Insect. Physiol. 2006, 52, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Mura, M.E.; Ruiu, L. Brevibacillus laterosporus pathogenesis and local immune response regulation in the house fly midgut. J. Invertebr. Pathol. 2017, 145, 55–61. [Google Scholar] [CrossRef]
- Kawasaki, K.; Andoh, M. Properties of induced antimicrobial activity in Musca domestica larvae. Drug Discov. Ther. 2017, 11, 156–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mourier, T.; Jeffares, D.C. Eukaryotic intron loss. Science 2003, 300, 1393. [Google Scholar] [CrossRef] [Green Version]
- Hogg, P.J. Disulfide bonds as switches for protein function. Trends Biochem. Sci. 2003, 28, 210–214. [Google Scholar] [CrossRef]
- Zhu, S.; Gao, B. Molecular characterization of a new scorpion venom lipolysis activating peptide: Evidence for disulfide bridge-mediated functional switch of peptides. FEBS Lett. 2006, 580, 6825–6836. [Google Scholar] [CrossRef] [PubMed] [Green Version]













| Model | l | LRT | Parameters | Positive Selected Sites |
|---|---|---|---|---|
| M0 | –918.761 | ω = 0.16294 | None | |
| M1a | –891.66 | ρ0 = 0.24997, ω0 = 0.00000 | Not allowed | |
| ρ1 = 0.75003, ω1 = 1.00000 | ||||
| M2a | –888.78 | 5.74 | ρ0 = 0.24996, ω0 = 0.00000 | |
| ρ1 = 0.66063, ω1 = 1.00000 | 7 S *, 22 L | |||
| ρ2 = 0.08941 ω2 = 3.89787 | ||||
| M7 | –875.388 | p = 0.39543, q = 1.05516 | Not allowed | |
| M8 | –875.26 | 0.26 | p0 = 0.97856, p = 0.41043, q = 1.16670 | 7 S |
| (p1 = 0.02144), ω = 2.30984 |
| Model | l | LRT | Parameters | Positive Selected Sites |
|---|---|---|---|---|
| M0 | –5115.94 | ω = 0.10448 | None | |
| M1a | –4880.09 | ρ0 = 0.32678, ω0 = 0.05181 | Not allowed | |
| ρ1 = 0.67322, ω1 = 1.00000 | ||||
| M2a | –4872.92 | 11.22 | ρ0 = 0.32607, ω0 = 0.05760 | 7R, 12K *, 14V, 18L *, 20P, 33E, 47P, 57Q |
| ρ1 = 0.52462, ω1 = 1.00000 | ||||
| ρ2 = 0.14931, ω2 = 2.14278 | ||||
| M7 | –4765.64 | p = 0.52104, q = 2.05131 | Not allowed | |
| M8 | –4765.64 | 0.00 | p0 = 1.00000, p = 0.52104, q = 2.05131 | None |
| (p1 = 0.00000), ω = 1.00000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, S.; Gao, B.; Zhu, S. Molecular Diversity and Evolution of Antimicrobial Peptides in Musca domestica. Diversity 2021, 13, 107. https://doi.org/10.3390/d13030107
Qi S, Gao B, Zhu S. Molecular Diversity and Evolution of Antimicrobial Peptides in Musca domestica. Diversity. 2021; 13(3):107. https://doi.org/10.3390/d13030107
Chicago/Turabian StyleQi, Sudong, Bin Gao, and Shunyi Zhu. 2021. "Molecular Diversity and Evolution of Antimicrobial Peptides in Musca domestica" Diversity 13, no. 3: 107. https://doi.org/10.3390/d13030107
APA StyleQi, S., Gao, B., & Zhu, S. (2021). Molecular Diversity and Evolution of Antimicrobial Peptides in Musca domestica. Diversity, 13(3), 107. https://doi.org/10.3390/d13030107