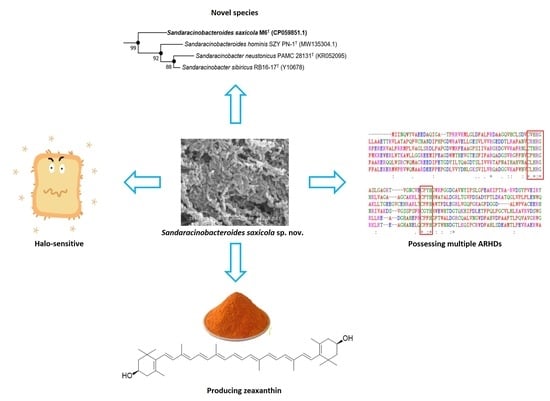
Sandaracinobacteroides saxicola sp. nov., a Zeaxanthin-Producing and Halo-Sensitive Bacterium Isolated from Fully Weathered Granitic Soil, and the Diversity of Its ARHDs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Maintenance of the Organisms
2.2. Phenotypic and Biochemical Characteristics
2.3. Chemotaxonomic Analysis
2.4. The 16S rRNA Gene Sequencing and Phylogenetic Analysis
2.5. Complete Genome Sequencing and Phylogenomic Analysis
3. Results and Discussion
3.1. Phylogenetic Placement and Phylogenomics
3.2. Morphology and Metabolic Profile
3.3. Chemotaxonomic Characteristics
3.4. The Biosynthesis of Zeaxanthin
3.5. Multiple Copies of Aromatic Ring-Hydroxylating Dioxygenases
3.6. Description of Sandaracinobacteroides saxicola sp. nov.
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Qu, P.H.; Luo, H.M. Sandaracinobacteroides hominis gen. nov., sp. nov., isolated from human skin. Arch. Microbiol. 2021, 203, 5067–5074. [Google Scholar] [CrossRef] [PubMed]
- Yurkov, V.; Stackebrandt, E. Reorganization of the genus Erythromicrobium: Description of “Erythromicrobium sibiricum” as Sandaracinobacter sibiricus gen. nov., sp. nov., and of “Erythromicrobium ursincola” as Erythromonas ursincola gen. nov., sp. nov. Int. J. Syst. Bacteriol. 1997, 47, 1172–1178. [Google Scholar] [CrossRef]
- Lee, I.; Jang, I.G. Sandaracinobacter neustonicus sp. nov., isolated from the sea surface microlayer in the southwestern pacific ocean, and emended description of the genus Sandaracinobacter. Int. J. Syst. Evol. Microbiol. 2020, 70, 4698–4703. [Google Scholar] [CrossRef] [PubMed]
- Parte, A.C.; Sardà Carbasse, J. List of prokaryotic names with standing in nomenclature (LPSN) moves to the dsmz. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef] [PubMed]
- Overmann, J.; Abt, B. Presence and future of culturing bacteria. Annu. Rev. Microbiol. 2017, 71, 711–730. [Google Scholar] [CrossRef]
- Gueidan, C.; Villaseñor, C.R. A rock-inhabiting ancestor for mutualistic and pathogen-rich fungal lineages. Stud. Mycol. 2008, 61, 111–119. [Google Scholar] [CrossRef]
- Pascual, J.; Wüst, P.K. Novel isolates double the number of chemotrophic species and allow the first description of higher taxa in Acidobacteria subdivision 4. Syst. Appl. Microbiol. 2015, 38, 534–544. [Google Scholar] [CrossRef]
- Maltman, C.; Yurkov, V. The effect of tellurite on highly resistant freshwater aerobic anoxygenic phototrophs and their strategies for reduction. Microorganisms 2015, 3, 826–838. [Google Scholar] [CrossRef]
- Yurkov, V.V.; Krieger, S. Citromicrobium bathyomarinum, a novel aerobic bacterium isolated from deep-sea hydrothermal vent plume waters that contains photosynthetic pigment-protein complexes. J. Bacteriol. 1999, 181, 4517–4525. [Google Scholar] [CrossRef]
- Kim, M.; Oh, H.S. Towards a taxonomic coherence between average nucleotide identity and 16s rrna gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef]
- Yurkov, V.; Gorlenko, V. Erythrobacter sibiricus sp. nov., a new fresh-water aerobic bacterial species containing bacteriochlorophyll a. Microbiology 1990, 59, 85–89. [Google Scholar]
- Smibert, R.M.; Krieg, N.R. Phenotypic characterization. In Methods for General Molecular Bacteriology; Gerhadt, P., Murray, R.G.E., Eds.; American Society for Microbiology: Washington, DC, USA, 1994; pp. 607–654. [Google Scholar]
- Kovacs, N. Identification of Pseudomonas pyocyanea by the oxidase reaction. Nature 1956, 178, 703. [Google Scholar] [CrossRef] [PubMed]
- Minnikin, D.E.; O’Donnell, A.G. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 1984, 2, 233–241. [Google Scholar] [CrossRef]
- Shin, Y.K.; Lee, J.S. Isoprenoid quinone profiles of the Leclercia adecarboxylata KCTC 1036T. J. Microbiol. Biotechnol. 1996, 6, 68–69. [Google Scholar]
- Frank, J.A.; Reich, C.I. Critical evaluation of two primers commonly used for amplification of bacterial 16s rRNA genes. Appl. Env. Microbiol. 2008, 74, 2461–2470. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M. Introducing ezbiocloud: A taxonomically united database of 16s rrna gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal omega, accurate alignment of very large numbers of sequences. Methods Mol. Biol. 2014, 1079, 105–116. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Rzhetsky, A.; Nei, M. A simple method for estimating and testing minimum-evolution trees. Mol. Biol. Evol. 1992, 9, 945–967. [Google Scholar]
- Kumar, S.; Stecher, G. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M.J. A simple method for estimating evolutionary rate of base substitution through comparative studies of nucleotide sequences. J. Mol. Evol. 1979, 16, 111–120. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limit on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; David, R. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Meier-Kolthoff, J.P.; Auch, A.F. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.M. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef]
- Avram, O.; Rapoport, D. M1CR0B1aAL1Z3R-a user-friendly web server for the analysis of large-scale microbial genomics data. Nucleic Acids Res. 2019, 47, W88–W92. [Google Scholar] [CrossRef]
- Alanjary, M.; Steinke, K. Automlst: An automated web server for generating multi-locus species trees highlighting natural product potential. Nucleic Acids Res. 2019, 47, W276–W282. [Google Scholar] [CrossRef]
- Qin, Q.L.; Xie, B.B. A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 2014, 196, 2210–2215. [Google Scholar] [CrossRef]
- Rodriguez-R, L.M.; Konstantinidis, K.T. Bypassing cultivation to identify bacterial species. Microbe 2014, 9, 111–118. [Google Scholar] [CrossRef]
- Yarza, P.; Richter, M. The all-species living tree project: A 16s rRNA-based phylogenetic tree of all sequenced type strains. Syst. Appl. Microbiol. 2008, 31, 241–250. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
- Alikhan, N.F.; Petty, N.K. Blast ring image generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [PubMed]
- Ohta, H.; Hattori, R. Sphingomonas oligophenolica sp. nov., a halo- and organo-sensitive oligotrophic bacterium from paddy soil that degrades phenolic acids at low concentrations. Int. J. Syst. Evol. Microbiol. 2004, 54, 2185–2190. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S. Antismash 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Shen, W. Darhd: A sequence database for aromatic ring-hydroxylating dioxygenase analysis and primer evaluation. J. Hazard Mater. 2022, 436, 129230. [Google Scholar] [CrossRef]
- Nojiri, H.; Ashikawa, Y. Structure of the terminal oxygenase component of angular dioxygenase, carbazole 1,9a-dioxygenase. J. Mol. Biol. 2005, 351, 355–370. [Google Scholar]
- Botelho, H.M.; Leal, S.S. Role of a novel disulfide bridge within the all-beta fold of soluble rieske proteins. J. Biol. Inorg. Chem. 2010, 15, 271–281. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J. Rasttk: A modular and extensible implementation of the rast algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [Green Version]







| Strains Pair | 16S% Identity | POCP Value | AAI | dDDH | ANI |
|---|---|---|---|---|---|
| M6T vs. Sandaracinobacteroides hominis | 96.3% | 80.7% | 62.1% | 18.0% | 72.6% |
| M6T vs. Sandaracinobacter neustonicus | 95.1% | 79.2% | 62.4% | 18.4% | 72.9% |
| S. hominis vs. S. neustonicus | 96.6% | 88.2% | 74.2% | 21.1% | 77.2% |
| Characteristics | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Oxygen requirement | Aerobic | Obligately aerobic | Facultatively anaerobic | Strictly aerobic |
| Growth at/with: | ||||
| NaCl (%, w/v) | 0 | 0–1.0 | 0.5–1.0 | 0–1.0 |
| Temperature (optimum) (°C) | 15–37 (30) | 10–37 (30) | 4–37 (30) | (25–30) |
| pH range (optimum) | 6.0–9.0 (6.0–8.5) | 6.0–8.0 (7.0) | 6.0–8.0 (6.5–7.0) | (7.5–8.5) |
| Motility | + | − | − | + |
| Bacteriochlorophyll a | − | − | − | + |
| Major carotenoid peaks (nm) | 450, 474 | 452, 478 | 450, 474 | 424, 450, 474 |
| Catalase activity | + | + | + | − |
| Oxidase activity | − | − | + | + |
| Hydrolysis of: | ||||
| Starch | + | + | + | − |
| Tweens (80) | − | − | + | ND |
| Quinone(s) | Q-9, Q-10 | Q-10, Q-11 | Q-10 | Q-9, Q-10 |
| DNA G+C content (mol %) | 67.7% | 65.0% | 65.3% | 68.5% * |
| Enzyme activities: | ||||
| Esterase lipase (C8) | − | weakly | − | ND |
| Esterase (C4) | − | + | − | ND |
| Valine arylamidase | + | − | ND | ND |
| Cystine arylamidase | + | − | ND | ND |
| α-chymotrypsin | − | weakly | + | ND |
| α-Galactosidase | + | − | − | ND |
| β-Galactosidase | + | − | + | ND |
| Acid production from: | ||||
| Aesculin | + | + | − | ND |
| d-Maltose | − | − | + | ND |
| Potassium 5-ketogluconate | − | − | + | ND |
| Main polar lipids | PG, PE, PL1–4, GL, L1-4 | DPG, PE, PG, SGL1-2, GL1-4, L1-7 | PG, PE, PL1–2, AL, GL, L | ND |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, Y.; Zhang, C.; Long, Q.; Lei, P.; Guo, Z.; Liu, Q. Sandaracinobacteroides saxicola sp. nov., a Zeaxanthin-Producing and Halo-Sensitive Bacterium Isolated from Fully Weathered Granitic Soil, and the Diversity of Its ARHDs. Diversity 2022, 14, 807. https://doi.org/10.3390/d14100807
Tang Y, Zhang C, Long Q, Lei P, Guo Z, Liu Q. Sandaracinobacteroides saxicola sp. nov., a Zeaxanthin-Producing and Halo-Sensitive Bacterium Isolated from Fully Weathered Granitic Soil, and the Diversity of Its ARHDs. Diversity. 2022; 14(10):807. https://doi.org/10.3390/d14100807
Chicago/Turabian StyleTang, Ying, Cuiyang Zhang, Qingshan Long, Ping Lei, Zhaohui Guo, and Qingshu Liu. 2022. "Sandaracinobacteroides saxicola sp. nov., a Zeaxanthin-Producing and Halo-Sensitive Bacterium Isolated from Fully Weathered Granitic Soil, and the Diversity of Its ARHDs" Diversity 14, no. 10: 807. https://doi.org/10.3390/d14100807
APA StyleTang, Y., Zhang, C., Long, Q., Lei, P., Guo, Z., & Liu, Q. (2022). Sandaracinobacteroides saxicola sp. nov., a Zeaxanthin-Producing and Halo-Sensitive Bacterium Isolated from Fully Weathered Granitic Soil, and the Diversity of Its ARHDs. Diversity, 14(10), 807. https://doi.org/10.3390/d14100807