The UDP-Glycosyltransferase Gene Family in Achelura yunnanensis (Lepidoptera: Zygaenidae): Identification, Phylogeny, and Diverse Expression Patterns
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insects and Tissues
2.2. RNA Extraction and Synthesis of First-Strand cDNA
2.3. Identification of Candidate UGTs in A. yunnanensis
2.4. Sequence and Phylogenetic Analysis of UGTs
2.5. Expression Profiling Analysis of UGT Genes in A. yunnanensis
3. Results
3.1. Identification of A. yunnanensis UGT Genes
3.2. Sequence and Phylogenetic Characteristics of A. yunnanensis UGTs
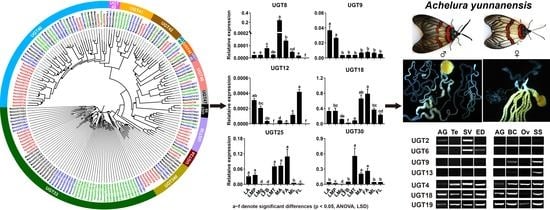
3.3. Stage- and Sex-Specific Expression Profile of A. yunnanensis UGT Genes
3.4. Candidate UGT Genes in A. yunnanensis Involving Reproduction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Heidel-Fischer, H.M.; Vogel, H. Molecular mechanisms of insect adaptation to plant secondary compounds. Curr. Opin. Insect Sci. 2015, 8, 8–14. [Google Scholar] [CrossRef]
- Despres, L.; David, J.P.; Gallet, C. The evolutionary ecology of insect resistance to plant chemicals. Trends Ecol. Evol. 2007, 22, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.-J.; Chertemps, T.; Maïbèche, M.; Marygold, S.J.; Van Leeuwen, T. Editorial: Invertebrate UDP-Glycosyltransferases: Nomenclature, Diversity and Functions. Front. Physiol. 2021, 12, 748290. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.J.; Vogel, H.; Heckel, D.G. Comparative analysis of the UDP-glycosyltransferase multigene family in insects. Insect Biochem. Mol. Biol. 2012, 42, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Nagare, M.; Ayachit, M.; Agnihotri, A.; Schwab, W.; Joshi, R. Glycosyltransferases: The multifaceted enzymatic regulator in insects. Insect Mol. Biol. 2021, 30, 123–137. [Google Scholar] [CrossRef]
- Heppner, J.B. Butterflies and Moths (Lepidoptera). In Encyclopedia of Entomology; Capinera, J.L., Ed.; Springer: Dordrecht, The Netherlands, 2008; pp. 626–672. [Google Scholar]
- Mitter, C.; Davis, D.R.; Cummings, M.P. Phylogeny and evolution of Lepidoptera. Annu. Rev. Entomol. 2017, 62, 265–283. [Google Scholar] [CrossRef]
- Bozzolan, F.; Siaussat, D.; Maria, A.; Durand, N.; Pottier, M.A.; Chertemps, T.; Maibeche-Coisne, M. Antennal uridine diphosphate (UDP)-glycosyltransferases in a pest insect: Diversity and putative function in odorant and xenobiotics clearance. Insect Mol. Biol. 2014, 23, 539–549. [Google Scholar] [CrossRef]
- Robertson, H.M.; Martos, R.; Sears, C.R.; Todres, E.Z.; Walden, K.K.; Nardi, J.B. Diversity of odourant binding proteins revealed by an expressed sequence tag project on male Manduca sexta moth antennae. Insect Mol. Biol. 1999, 8, 501–518. [Google Scholar] [CrossRef]
- Bock, K.W. Vertebrate UDP-glucuronosyltransferases: Functional and evolutionary aspects. Biochem. Pharmacol. 2003, 66, 691–696. [Google Scholar] [CrossRef]
- Huang, F.F.; Chai, C.L.; Zhang, Z.; Liu, Z.H.; Dai, F.Y.; Lu, C.; Xiang, Z.H. The UDP-glucosyltransferase multigene family in Bombyx mori. BMC Genom. 2008, 9, 563. [Google Scholar] [CrossRef] [Green Version]
- Luque, T.; Okano, K.; O’Reilly, D.R. Characterization of a novel silkworm (Bombyx mori) phenol UDP-glucosyltransferase. Eur. J. Biochem. 2002, 269, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Daimon, T.; Hirayama, C.; Kanai, M.; Ruike, Y.; Meng, Y.; Kosegawa, E.; Nakamura, M.; Tsujimoto, G.; Katsuma, S.; Shimada, T. The silkworm Green b locus encodes a quercetin 5-O-glucosyltransferase that produces green cocoons with UV-shielding properties. Proc. Natl. Acad. Sci. USA 2010, 107, 11471–11476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krempl, C.; Sporer, T.; Reichelt, M.; Ahn, S.-J.; Heidel-Fischer, H.; Vogel, H.; Heckel, D.G.; Joußen, N. Potential detoxification of gossypol by UDP-glycosyltransferases in the two Heliothine moth species Helicoverpa armigera and Heliothis virescens. Insect Biochem. Mol. Biol. 2016, 71, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Israni, B.; Wouters, F.C.; Luck, K.; Seibel, E.; Ahn, S.-J.; Paetz, C.; Reinert, M.; Vogel, H.; Erb, M.; Heckel, D.G.; et al. The fall armyworm Spodoptera frugiperda utilizes specific UDP-glycosyltransferases to inactivate maize defensive benzoxazinoids. Front. Physiol. 2020, 11, 604754. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.J.; Badenes-Perez, F.R.; Reichelt, M.; Svatos, A.; Schneider, B.; Gershenzon, J.; Heckel, D.G. Metabolic detoxification of capsaicin by UDP-glycosyltransferase in three Helicoverpa species. Arch. Insect. Biochem. Physiol. 2011, 78, 104–118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-N.; Ma, J.-F.; Xu, L.; Dong, Z.-P.; Xu, J.-W.; Li, M.-Y.; Zhu, X.-Y. Identification and expression patterns of UDP-glycosyltransferase (UGT) genes from insect pest Athetis lepigone (Lepidoptera: Noctuidae). J. Asia-Pac. Entomol. 2017, 20, 253–259. [Google Scholar] [CrossRef]
- He, P.; Zhang, Y.-F.; Hong, D.-Y.; Wang, J.; Wang, X.-L.; Zuo, L.-H.; Tang, X.-F.; Xu, W.-M.; He, M. A reference gene set for sex pheromone biosynthesis and degradation genes from the diamondback moth, Plutella xylostella, based on genome and transcriptome digital gene expression analyses. BMC Genom. 2017, 18, 219. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Zhang, S.H.; Ren, M.M.; Tian, X.R.; Wei, Q.; Mburu, D.K.; Su, J.Y. The expression of Spodoptera exigua P450 and UGT genes: Tissue specificity and response to insecticides. Insect Sci. 2019, 26, 199–216. [Google Scholar] [CrossRef] [Green Version]
- Li, G.-C.; Nuo, S.-M.; Wang, Z.-Q.; Yang, A.-J.; Liu, N.-Y. Identification and expression profiling of chemosensory membrane protein genes in Achelura yunnanensis (Lepidoptera: Zygaenidae). Comp. Biochem. Physiol. D Genom. Proteom. 2021, 40, 100876. [Google Scholar] [CrossRef]
- Nuo, S.M.; Yang, A.J.; Li, G.C.; Xiao, H.Y.; Liu, N.Y. Transcriptome analysis identifies candidate genes in the biosynthetic pathway of sex pheromones from a zygaenid moth, Achelura yunnanensis (Lepidoptera: Zygaenidae). PeerJ 2021, 9, e12641. [Google Scholar] [CrossRef]
- Li, G.C.; Zhao, Y.J.; Li, J.L.; Lu, G.Y.; Liu, N.Y. Ultrastructure of sensilla on the antennae, proboscis and tarsi of adult Achelura yunnanensis (Lepidoptera: Zygaenidae). Acta Entomol. Sin. 2020, 63, 1385–1398. [Google Scholar]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.; Zhou, Z.; Lu, C.; Cheng, D.; Dai, F.; Li, B.; Zhao, P.; Zha, X.; Cheng, T.; Chai, C.; et al. A draft sequence for the genome of the domesticated silkworm Bombyx mori. Science 2004, 306, 1937. [Google Scholar]
- Kanost, M.R.; Arrese, E.L.; Cao, X.; Chen, Y.R.; Chellapilla, S.; Goldsmith, M.R.; Grosse-Wilde, E.; Heckel, D.G.; Herndon, N.; Jiang, H.; et al. Multifaceted biological insights from a draft genome sequence of the tobacco hornworm moth, Manduca sexta. Insect Biochem. Mol. Biol. 2016, 76, 118–147. [Google Scholar] [CrossRef]
- Pearce, S.L.; Clarke, D.F.; East, P.D.; Elfekih, S.; Gordon, K.H.J.; Jermiin, L.S.; McGaughran, A.; Oakeshott, J.G.; Papanikolaou, A.; Perera, O.P.; et al. Genomic innovations, transcriptional plasticity and gene loss underlying the evolution and divergence of two highly polyphagous and invasive Helicoverpa pest species. BMC Biol. 2017, 15, 63. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, P. Q-Gene: Processing quantitative real-time RT-PCR data. Bioinformatics 2003, 19, 1439–1440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muller, P.Y.; Janovjak, H.; Miserez, A.R.; Dobbie, Z. Processing of gene expression data generated by quantitative real-time RT-PCR. BioTechniques 2002, 32, 1372–1374, 1376, 1378–1379. [Google Scholar] [PubMed]
- Miley, M.J.; Zielinska, A.K.; Keenan, J.E.; Bratton, S.M.; Radominska-Pandya, A.; Redinbo, M.R. Crystal structure of the cofactor-binding domain of the human phase II drug-metabolism enzyme UDP-glucuronosyltransferase 2B7. J. Mol. Biol. 2007, 369, 498–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davidson, N.M.; Oshlack, A. Corset: Enabling differential gene expression analysis for de novo assembled transcriptomes. Genome Biol. 2014, 15, 410. [Google Scholar] [PubMed] [Green Version]
- Xiao, H.Y.; Li, G.C.; Wang, Z.Q.; Guo, Y.R.; Liu, N.Y. Combined transcriptomic, proteomic and genomic analysis identifies reproductive-related proteins and potential modulators of female behaviors in Spodoptera litura. Genom. 2021, 113, 1876–1894. [Google Scholar] [CrossRef] [PubMed]
- Swanson, W.J.; Clark, A.G.; Waldrip-Dail, H.M.; Wolfner, M.F.; Aquadro, C.F. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc. Natl. Acad. Sci. USA 2001, 98, 7375–7379. [Google Scholar] [CrossRef] [Green Version]
- Wilburn, D.B.; Swanson, W.J. From molecules to mating: Rapid evolution and biochemical studies of reproductive proteins. J. Proteom. 2016, 135, 12–25. [Google Scholar] [CrossRef] [Green Version]
- Celorio-Mancera, M.d.l.P.; Ahn, S.-J.; Vogel, H.; Heckel, D.G. Transcriptional responses underlying the hormetic and detrimental effects of the plant secondary metabolite gossypol on the generalist herbivore Helicoverpa armigera. BMC Genom. 2011, 12, 575. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Zhu, B.; Gao, X.; Liang, P. Over-expression of UDP–glycosyltransferase gene UGT2B17 is involved in chlorantraniliprole resistance in Plutella xylostella (L.). Pest Manag. Sci. 2017, 73, 1402–1409. [Google Scholar] [CrossRef]
- Yin, N.-N.; Zhao, Y.-J.; Zhu, J.-Y.; Liu, N.-Y. Antennal UDP-glycosyltransferase genes in the coffee white stemborer, Xylotrechus quadripes. J. Asia-Pac. Entomol. 2019, 22, 1145–1153. [Google Scholar] [CrossRef]
- Wang, Z.Q.; Yin, N.N.; Zhao, N.; Liu, N.Y. Identification and expression characterization of UDP-glucosyltransferase genes in Rhaphuma horsfieldi. Chinese J. Appl. Entomol. 2020, 57, 898–910. [Google Scholar]
- McKenna, D.D.; Scully, E.D.; Pauchet, Y.; Hoover, K.; Kirsch, R.; Geib, S.M.; Mitchell, R.F.; Waterhouse, R.M.; Ahn, S.J.; Arsala, D.; et al. Genome of the Asian longhorned beetle (Anoplophora glabripennis), a globally significant invasive species, reveals key functional and evolutionary innovations at the beetle-plant interface. Genome Biol. 2016, 17, 227. [Google Scholar] [CrossRef] [Green Version]
- Gouin, A.; Bretaudeau, A.; Nam, K.; Gimenez, S.; Aury, J.-M.; Duvic, B.; Hilliou, F.; Durand, N.; Montagné, N.; Darboux, I.; et al. Two genomes of highly polyphagous lepidopteran pests (Spodoptera frugiperda, Noctuidae) with different host-plant ranges. Sci. Rep. 2017, 7, 11816. [Google Scholar] [CrossRef] [PubMed]
- Zagrobelny, M.; Scheibye-Alsing, K.; Jensen, N.B.; Møller, B.L.; Gorodkin, J.; Bak, S. 454 pyrosequencing based transcriptome analysis of Zygaena filipendulae with focus on genes involved in biosynthesis of cyanogenic glucosides. BMC Genom. 2009, 10, 574. [Google Scholar] [CrossRef] [Green Version]
- Thrimawithana, A.H.; Wu, C.; Christeller, J.T.; Simpson, R.M.; Hilario, E.; Tooman, L.K.; Begum, D.; Jordan, M.D.; Crowhurst, R.; Newcomb, R.D.; et al. The genomics and population genomics of the light brown apple moth, Epiphyas postvittana, an invasive tortricid pest of horticulture. Insects 2022, 13, 264. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Shi, H.; Gao, X.; Liang, P. Characterization of UDP-glucuronosyltransferase genes and their possible roles in multi-insecticide resistance in Plutella xylostella (L.). Pest Manag. Sci. 2018, 74, 695–704. [Google Scholar] [CrossRef]
- Jensen, N.B.; Zagrobelny, M.; Hjernø, K.; Olsen, C.E.; Houghton-Larsen, J.; Borch, J.; Møller, B.L.; Bak, S. Convergent evolution in biosynthesis of cyanogenic defence compounds in plants and insects. Nat. Commun. 2011, 2, 273. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.Z.; Gong, W.C.; Ma, Q.Y.; Niu, Y.; Mei, W.L.; Chen, C.; Zhao, Y.X. Isolation and identification of cyanogenic glycosides from larval secretions of Achelura yunnanensis ( Lepidoptera: Zygaenidae) and the bioactivity against Tapinoma melanocephalum (Hymenoptera: Fromicidae). Acta Entomol. Sin. 2013, 56, 207–211. [Google Scholar]
- Wang, Q.; Hasan, G.; Pikielny, C.W. Preferential expression of biotransformation enzymes in the olfactory organs of Drosophila melanogaster, the antennae. J. Biol. Chem. 1999, 274, 10309–10315. [Google Scholar] [CrossRef] [Green Version]
- Younus, F.; Chertemps, T.; Pearce, S.L.; Pandey, G.; Bozzolan, F.; Coppin, C.W.; Russell, R.J.; Maibeche-Coisne, M.; Oakeshott, J.G. Identification of candidate odorant degrading gene/enzyme systems in the antennal transcriptome of Drosophila melanogaster. Insect Biochem. Mol. Biol. 2014, 53, 30–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fraichard, S.; Legendre, A.; Lucas, P.; Chauvel, I.; Faure, P.; Neiers, F.; Artur, Y.; Briand, L.; Ferveur, J.-F.; Heydel, J.-M. Modulation of sex pheromone discrimination by a UDP-glycosyltransferase in Drosophila melanogaster. Genes 2020, 11, 237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Liu, Y.; Zhou, J.J.; Yi, J.K.; Pan, Y.; Wang, J.; Zhang, X.X.; Wang, J.X.; Yang, S.; Xi, J.H. Identification and tissue expression profiling of candidate UDP-glycosyltransferase genes expressed in Holotrichia parallela motschulsky antennae. Bull. Entomol. Res. 2018, 108, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.-J.; Marygold, S.J. The UDP-glycosyltransferase family in Drosophila melanogaster: Nomenclature update, gene expression and phylogenetic analysis. Front. Physiol. 2021, 12, 648481. [Google Scholar] [CrossRef] [PubMed]






| Gene | ORF (bp) | SP (aa) | pI | Mw (kDa) | NPS |
|---|---|---|---|---|---|
| UGT1 | 1560 | 18 | 7.73 | 59.90 | 114, 273, 419 |
| UGT2 | 1557 | 17 | 7.32 | 59.53 | 63, 330 |
| UGT3 | 1593 | 17 | 8.89 | 60.03 | 63, 184, 247, 294, 343 |
| UGT4 | 1575 | 18 | 7.65 | 60.28 | 67, 427, 474 |
| UGT5 | 1542 | 18 | 8.78 | 58.27 | 49, 169, 318, 447, 501 |
| UGT6 | 1602 | 19 | 7.68 | 61.39 | 68, 138, 199, 308, 479, 523 |
| UGT7 | 1575 | 21 | 6.52 | 61.29 | 52, 244, 326, 452, 516 |
| UGT8 | 1563 | 20 | 9.11 | 58.33 | 237, 302 |
| UGT9 | 1554 | 20 | 9.18 | 59.29 | 82, 175, 245, 511 |
| UGT10 | 1641 | 21 | 8.97 | 62.57 | 52, 239, 333 |
| UGT11 | 1545 | 17 | 9.22 | 59.21 | 71, 89, 102, 237, 285 |
| UGT12 | 1563 | 20 | 9.06 | 58.27 | 103, 237, 302 |
| UGT13 | 1557 | 20 | 8.95 | 59.75 | 128 |
| UGT14 | 1569 | 22 | 7.71 | 60.69 | 71, 192, 289, 472 |
| UGT15 | 1545 | 18 | 8.98 | 59.13 | 120, 240 |
| UGT16 | 1581 | 20 | 6.37 | 60.23 | 147, 236 |
| UGT17 | 1566 | 20 | 8.97 | 60.88 | 51, 93, 240, 274, 426, 449 |
| UGT18 | 1593 | 21 | 8.83 | 60.56 | 252, 436 |
| UGT19 | 1539 | 19 | 8.84 | 59.41 | 60, 65, 102 |
| UGT20 | 1563 | 18 | 7.29 | 59.64 | 117, 251 |
| UGT21 | 1512 | 15 | 7.34 | 57.08 | 61, 72, 279, 315, 416, 417 |
| UGT22 | 1575 | 24 | 7.25 | 60.83 | 73, 291 |
| UGT23 | 1584 | 20 | 6.55 | 60.81 | 69, 429, 477 |
| UGT24 | 1536 | 21 | 8.99 | 58.60 | 74, 238, 289 |
| UGT25 | 1554 | 17 | 8.10 | 59.96 | 66, 147, 463 |
| UGT26 | 1578 | 22 | 7.59 | 60.24 | 71, 477 |
| UGT27 | 1557 | 17 | 6.69 | 59.51 | 66, 189, 474, 515 |
| UGT28 | 1578 | 20 | 9.13 | 60.10 | 82, 175, 245 |
| UGT29 | 1623 | 16 | 9.50 | 61.29 | 78, 119, 205, 231, 327, 406 |
| UGT30 | 1539 | 19 | 8.76 | 59.34 | 65 |
| UGT31 | 1548 | 18 | 8.16 | 58.91 | 120, 240, 415, 507 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, H.-Y.; Chen, D.-L.; Lu, T.-T.; Yao, Y.-J.; Liu, N.-Y. The UDP-Glycosyltransferase Gene Family in Achelura yunnanensis (Lepidoptera: Zygaenidae): Identification, Phylogeny, and Diverse Expression Patterns. Diversity 2022, 14, 407. https://doi.org/10.3390/d14050407
Xiao H-Y, Chen D-L, Lu T-T, Yao Y-J, Liu N-Y. The UDP-Glycosyltransferase Gene Family in Achelura yunnanensis (Lepidoptera: Zygaenidae): Identification, Phylogeny, and Diverse Expression Patterns. Diversity. 2022; 14(5):407. https://doi.org/10.3390/d14050407
Chicago/Turabian StyleXiao, Hai-Yan, Dan-Lu Chen, Ting-Ting Lu, Yu-Juan Yao, and Nai-Yong Liu. 2022. "The UDP-Glycosyltransferase Gene Family in Achelura yunnanensis (Lepidoptera: Zygaenidae): Identification, Phylogeny, and Diverse Expression Patterns" Diversity 14, no. 5: 407. https://doi.org/10.3390/d14050407
APA StyleXiao, H. -Y., Chen, D. -L., Lu, T. -T., Yao, Y. -J., & Liu, N. -Y. (2022). The UDP-Glycosyltransferase Gene Family in Achelura yunnanensis (Lepidoptera: Zygaenidae): Identification, Phylogeny, and Diverse Expression Patterns. Diversity, 14(5), 407. https://doi.org/10.3390/d14050407