Taxonomic Uncertainty and Its Conservation Implications in Management, a Case from Pyrus hopeiensis (Rosaceae)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Morphological Study
2.2. Phylogenomic Inference
3. Results
3.1. Morphological Study
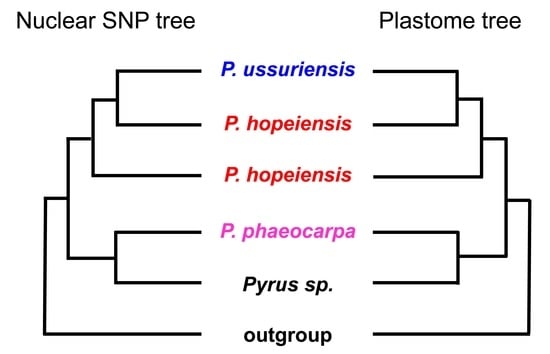
3.2. Phylogenomic Inference
4. Discussion
4.1. Integrative Evidence for Reappraisal of the Identity of Pyrus hopeiensis
4.2. Integrative Evidence-Based Taxonomy for Biodiversity Conservation
4.3. Taxonomic Treatment
Other Specimens Examined
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Cowie, R.H.; Bouchet, P.; Fontaine, B. The Sixth Mass Extinction: Fact, fiction or speculation? Biol. Rev. 2022, 97, 640–663. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Chen, G.; Edward Grumbine, R.; Dao, Z.; Sun, W.; Guo, H. Conserving plant species with extremely small populations (PSESP) in China. Biodivers. Conserv. 2013, 22, 803–809. [Google Scholar] [CrossRef]
- Lu, Z.; Qin, H.; Jin, X.; Zhang, Z.; Yang, Q.; Hong, D.; Li, D.; Li, K.; Yuan, L.; Zhou, Z. On the necessity, principle, and process of updating the List of National Key Protected Wild Plants. Biodivers. Sci. 2021, 29, 1577–1582. [Google Scholar] [CrossRef]
- Yang, Y.; Tan, C.; Yang, Z. Conservation of gymnosperms in China: Perspectives from the List of National Key Protected Wild Plants. Biodivers. Sci. 2021, 29, 1591–1598. [Google Scholar] [CrossRef]
- Heywood, V.H.; Davis, P.H. Principles of Angiosperm Taxonomy; Oliver and Boyd: Edinburgh, UK; London, UK, 1963. [Google Scholar]
- Dubois, A. Phylogeny, taxonomy, and nomenclature: The problem of taxonomic categories and nomenclatural ranks. Zootaxa 2007, 1519, 27–58. [Google Scholar] [CrossRef]
- Morrison, W.R.; Lohr, J.L.; Duchen, P.; Wilches, R.; Trujillo, D.; Mair, M.; Renner, S.S. The impact of taxonomic change on conservation: Does it kill, can it save, or is it just irrelevant? Biol. Conserv. 2009, 142, 3201–3206. [Google Scholar] [CrossRef]
- Goodwin, Z.; Harris, D.J.; Filer, D.; Wood, J.R.I.; Scotland, R.W. Widespread mistaken identity in tropical plant collections. Curr. Biol. 2015, 25, R1066–R1067. [Google Scholar] [CrossRef] [Green Version]
- Lian, L.; Shen, J.Y.; Ortiz, R.D.C.; Yu, S.X.; Chen, Z.D.; Wang, W. Phylogenetic analyses and modelling distributions guide conservation of a critically endangered liana species, Eleutharrhena macrocarpa (Menispermaceae). Taxon 2021, 70, 931–945. [Google Scholar] [CrossRef]
- Zhang, M.; Xue, C.; Hu, H.; Li, J.; Xue, Y.; Wang, R.; Fan, J.; Zou, C.; Tao, S.; Qin, M.; et al. Genome-wide association studies provide insights into the genetic determination of fruit traits of pear. Nat. Commun. 2021, 12, 1144. [Google Scholar] [CrossRef]
- Rubtsov, G.A. Geographical distribution of the genus Pyrus and trends and factors in its evolution. Am. Nat. 1944, 78, 358–366. [Google Scholar] [CrossRef]
- Korotkova, N.; Parolly, G.; Khachatryan, A.; Ghulikyan, L.; Sargsyan, H.; Akopian, J.; Borsch, T.; Gruenstaeudl, M. Towards resolving the evolutionary history of Caucasian pears (Pyrus, Rosaceae)-Phylogenetic relationships, divergence times and leaf trait evolution. J. Syst. Evol. 2018, 56, 35–47. [Google Scholar] [CrossRef]
- Dong, X.; Wang, Z.; Tian, L.; Zhang, Y.; Qi, D.; Huo, H.; Xu, J.; Li, Z.; Liao, R.; Shi, M.; et al. De novo assembly of a wild pear (Pyrus betuleafolia) genome. Plant Biotechnol. J. 2020, 18, 581–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Browicz, K. Concept and chorology of the genus Pyrus L. Arboret. Kórnick. 1993, 38, 17–33. [Google Scholar]
- Robertson, K.R.; Rohrer, J.R.; Rohrer, J.B.; Smith, P.G. A synopsis of genera in subfamily Maloideae (Rosaceae). Syst. Bot. 1991, 16, 376–394. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Z.; Shi, Z.; Zhang, S.; Ming, R.; Zhu, S.; Khan, M.A.; Tao, S.; Korban, S.S.; Wang, H.; et al. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 2013, 23, 396–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Singh, J.; Qin, M.; Li, S.; Zhang, X.; Zhang, M.; Khan, A.; Zhang, S.; Wu, J. Development of an integrated 200K SNP genotyping array and application for genetic mapping, genome assembly improvement and genome wide association studies in pear (Pyrus). Plant Biotechnol. J. 2019, 17, 1582–1594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirasawa, K.; Itai, A.; Isobe, S. Chromosome-scale genome assembly of Japanese pear (Pyrus pyrifolia) variety ‘Nijisseiki’. DNA Res. 2021, 28, b1. [Google Scholar] [CrossRef]
- Zheng, X.; Cai, D.; Potter, D.; Postman, J.; Liu, J.; Teng, Y. Phylogeny and evolutionary histories of Pyrus L. revealed by phylogenetic trees and networks based on data from multiple DNA sequences. Mol. Phylogenet. Evol. 2014, 80, 54–65. [Google Scholar] [CrossRef]
- Feng, S.; Bai, M.; Rivas-González, I.; Li, C.; Liu, S.; Tong, Y.; Yang, H.; Chen, G.; Xie, D.; Sears, K.E.; et al. Incomplete lineage sorting and phenotypic evolution in marsupials. Cell 2022, 185, 1646–1660.e18. [Google Scholar] [CrossRef]
- Mu, X.Y.; Sun, M.; Yang, P.F.; Lin, Q.W. Unveiling the identity of wenwan walnuts and phylogenetic relationships of Asian Juglans species using restriction site-associated DNA-sequencing. Front. Plant Sci. 2017, 8, 1708. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.B.; Liu, G.N.; Hong, D.Y.; Wen, J. Eriobotrya belongs to Rhaphiolepis (Maleae, Rosaceae): Evidence from chloroplast genome and nuclear ribosomal DNA data. Front. Plant Sci. 2020, 10, 1731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yü, T.T.; Ku, T.C. Pyrus. In Flora Reipublicae Popularis Sinicae; Yü, T.T., Ed.; Science Press: Beijing, China, 1974; Volume 36, pp. 354–372. [Google Scholar]
- Wuyun, T.; Ma, T.; Uematsu, C.; Katayama, H. A phylogenetic network of wild Ussurian pears (Pyrus ussuriensis Maxim.) in China revealed by hypervariable regions of chloroplast DNA. Tree Genet. Genomes 2013, 9, 167–177. [Google Scholar] [CrossRef]
- Yü, T.T.; Kuan, K.C. Taxa nova Rosacearum Sinicarum (I). Acta Phytotaxon. Sin. 1963, 8, 202–236. [Google Scholar]
- Liang, T.T.; Ma, Y.; Guo, J.; Zang, D.K. Transcriptome sequencing and analysis of wild pear (Pyrus hopeiensis) using the Illumina Platform. Arab. J. Sci. Eng. 2016, 41, 45–53. [Google Scholar] [CrossRef]
- Zang, R.G.; Dong, M.; Li, J.Q.; Chen, X.Y.; Zeng, S.J.; Jiang, M.X.; Li, Z.Q.; Huang, J.H. Conservation and restoration for typical critically endangered wild plants with extremely small population. Acta Ecol. Sin. 2016, 36, 7130–7135. [Google Scholar] [CrossRef]
- Li, W.; Lu, Y.; Xie, X.; Li, B.; Han, Y.; Sun, T.; Xian, Y.; Yang, H.; Liu, K. Development of chloroplast genomic resources for Pyrus hopeiensis (Rosaceae). Conserv. Genet. Resour. 2018, 10, 511–513. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, J.; Li, L.; Gao, L.; Xu, J.; Yang, M. Structural and comparative analysis of the complete chloroplast genome of Pyrus hopeiensis “Wild Plants with a Tiny Population” and three other Pyrus species. Int. J. Mol. Sci. 2018, 19, 3262. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Tian, L.; Gao, Y.; Liu, F. Genetic diversity of cultivated and wild Ussurian Pear (Pyrus ussuriensis Maxim.) in China evaluated with M13-tailed SSR markers. Genet. Resour. Crop Evol. 2012, 59, 9–17. [Google Scholar] [CrossRef]
- Liu, Q.; Kang, M.; Jiang, Y. Some new recorded plants from Beijing and Hebei (II). J. Beijing Norm. Univ. 2003, 39, 674–676. [Google Scholar]
- Wang, Q. Identity of Microtoena coreana (Lamiaceae), a doubtful species from Korea. Phytotaxa 2015, 195, 79–85. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Y.; Xu, J.; Korban, S.S.; Fei, Z.; Tao, S.; Ming, R.; Tai, S.; Khan, A.M.; Postman, J.D.; et al. Diversification and independent domestication of Asian and European pears. Genome Biol. 2018, 19, 77. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, J.; Wang, S.; Zhang, Y.; Yang, M. The distribution and origins of Pyrus hopeiensis-“Wild Plant With Tiny Population” using whole genome resequencing. Front. Plant Sci. 2021, 12, 668796. [Google Scholar] [CrossRef] [PubMed]
- Velasco, R.; Zharkikh, A.; Affourtit, J.; Dhingra, A.; Cestaro, A.; Kalyanaraman, A.; Fontana, P.; Bhatnagar, S.K.; Troggio, M.; Pruss, D.; et al. The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 2010, 42, 833–839. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; DePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Qu, Y.; Wang, R.; Ma, C.; Li, D.; Song, J. Genetic diversity analysis of pear genus in Shanxi Province by SSR markers. North. Hort. 2015, 39, 11–17. [Google Scholar]
- Xiang, Y.; Huang, C.; Hu, Y.; Wen, J.; Li, S.; Yi, T.; Chen, H.; Xiang, J.; Ma, H. Evolution of Rosaceae fruit types based on nuclear phylogeny in the context of geological times and genome duplication. Mol. Biol. Evol. 2017, 34, 262–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.D.; Jin, J.J.; Chen, S.Y.; Chase, M.W.; Soltis, D.E.; Li, H.T.; Yang, J.B.; Li, D.Z.; Yi, T.S. Diversification of Rosaceae since the Late Cretaceous based on plastid phylogenomics. New Phytol. 2017, 214, 1355–1367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.B.; Ren, C.; Kwak, M.; Hodel, R.G.J.; Xu, C.; He, J.; Zhou, W.B.; Huang, C.H.; Ma, H.; Qian, G.Z.; et al. Phylogenomic conflict analyses in the apple genus Malus s.l. reveal widespread hybridization and allopolyploidy driving diversification, with insights into the complex biogeographic history in the Northern Hemisphere. J. Integr. Plant Biol. 2022, 64, 1020–1043. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Hu, C.; Spooner, D.; Liu, J.; Cao, J.; Teng, Y. Molecular evolution of Adh and LEAFY and the phylogenetic utility of their introns in Pyrus (Rosaceae). BMC Evol. Biol. 2011, 11, 255. [Google Scholar] [CrossRef] [Green Version]
- Corlett, R.T. Safeguarding our future by protecting biodiversity. Plant Divers. 2020, 42, 221–228. [Google Scholar] [CrossRef]
- Raven, P.; Wackernagel, M. Maintaining biodiversity will define our long-term success. Plant Divers. 2020, 42, 211–220. [Google Scholar] [CrossRef]
- Bremer, K.; Bremer, B.; Karis, P.; Kallersjo, M. Time for change in taxonomy. Nature 1990, 343, 202. [Google Scholar] [CrossRef]
- Raposo, M.A.; Stopiglia, R.; Brito, G.R.R.; Bockmann, F.A.; Kirwan, G.M.; Gayon, J.; Dubois, A. What really hampers taxonomy and conservation? A riposte to Garnett and Christidis (2017). Zootaxa 2017, 4317, 179–184. [Google Scholar] [CrossRef]
- Hu, H.H. Notulae systematicae ad Florem Sinensium V. Bull. Fan Mem. Inst. Biol. 1934, 5, 305–306. [Google Scholar]
- Zhang, W.P.; Cao, L.; Lin, X.R.; Ding, Y.M.; Liang, Y.; Zhang, D.Y.; Pang, E.L.; Renner, S.S.; Bai, W.N. Dead-end hybridization in walnut trees revealed by large-scale genomic sequence data. Mol. Biol. Evol. 2021, 39, msab308. [Google Scholar] [CrossRef]
- Krofel, M.; Hatlauf, J.; Bogdanowicz, W.; Campbell, L.A.D.; Godinho, R.; Jhala, Y.V.; Kitchener, A.C.; Koepfli, K.P.; Moehlman, P.; Senn, H.; et al. Towards resolving taxonomic uncertainties in wolf, dog and jackal lineages of Africa, Eurasia and Australasia. J. Zool. 2022, 316, 155–168. [Google Scholar] [CrossRef]
- Albani Rocchetti, G.; Armstrong, C.G.; Abeli, T.; Orsenigo, S.; Jasper, C.; Joly, S.; Bruneau, A.; Zytaruk, M.; Vamosi, J.C. Reversing extinction trends: New uses of (old) herbarium specimens to accelerate conservation action on threatened species. New Phytol. 2021, 230, 433–450. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-C., D.; Richardson, J.E.; Hart, M.; Serrano, J.; Cárdenas, D.; Gonzalez, M.A.; Cortés-B., R. A plea to DNA barcode type specimens: An example from Micropholis (Sapotaceae). Taxon 2022, 71, 154–167. [Google Scholar] [CrossRef]
- Hart, M.L.; Forrest, L.L.; Nicholls, J.A.; Kidner, C.A. Retrieval of hundreds of nuclear loci from herbarium specimens. Taxon 2016, 65, 1081–1092. [Google Scholar] [CrossRef]
- Silva, C.; Besnard, G.; Piot, A.; Razanatsoa, J.; Oliveira, R.P.; Vorontsova, M.S. Museomics resolve the systematics of an endangered grass lineage endemic to north-western Madagascar. Ann. Bot. 2017, 119, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Raxworthy, C.J.; Smith, B.T. Mining museums for historical DNA: Advances and challenges in museomics. Trends Ecol. Evol. 2021, 36, 1049–1060. [Google Scholar] [CrossRef]





| Specimens of the Flowering Period | Specimens of the Fruiting Period | ||
|---|---|---|---|
| 1 | Leaf blade length [cm] | 1 | Leaf blade length [cm] |
| 2 | Leaf blade width [cm] | 2 | Leaf blade width [cm] |
| 3 | Ratio of leaf blade width to length | 3 | Ratio of leaf blade width to length |
| 4 | Leaf serration: (1) long; (0) short | 4 | Leaf serration: (1) long; (0) short |
| 5 | Length of flower stalk [cm] | 5 | Length of fruit [cm] |
| 6 | Indumentum density on stalk: (0) sparse; (1) dense | 6 | Persisting calyx: (0) no; (1) yes |
| 7 | Length of pedicel [cm] | ||
| 8 | Ratio of fruit to pedicel Length | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mu, X.-Y.; Wu, J.; Wu, J. Taxonomic Uncertainty and Its Conservation Implications in Management, a Case from Pyrus hopeiensis (Rosaceae). Diversity 2022, 14, 417. https://doi.org/10.3390/d14060417
Mu X-Y, Wu J, Wu J. Taxonomic Uncertainty and Its Conservation Implications in Management, a Case from Pyrus hopeiensis (Rosaceae). Diversity. 2022; 14(6):417. https://doi.org/10.3390/d14060417
Chicago/Turabian StyleMu, Xian-Yun, Jiang Wu, and Jun Wu. 2022. "Taxonomic Uncertainty and Its Conservation Implications in Management, a Case from Pyrus hopeiensis (Rosaceae)" Diversity 14, no. 6: 417. https://doi.org/10.3390/d14060417
APA StyleMu, X. -Y., Wu, J., & Wu, J. (2022). Taxonomic Uncertainty and Its Conservation Implications in Management, a Case from Pyrus hopeiensis (Rosaceae). Diversity, 14(6), 417. https://doi.org/10.3390/d14060417