A Multichannel Fluorescent Array Sensor for Discrimination of Different Types of Drug-Induced Kidney Injury
Abstract
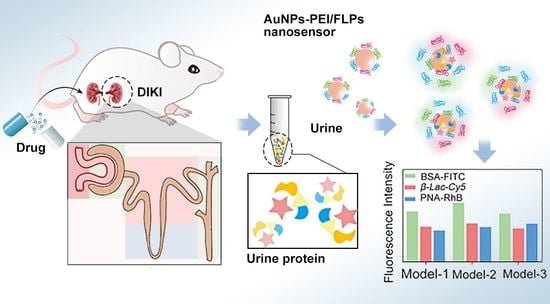
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Instruments
2.3. Preparing of AuNPs–PEI/FLPs Sensor
2.4. Fluorescence Titrations
2.5. Animals and Experimental Design
- (1)
- Normal control group. Mice received saline.
- (2)
- Aminoglycoside antibiotics (AGs) group. Gentamicin (97.07 mg/kg/d), tobramycin (46.41 mg/kg/d), neomycin (303 mg/kg/d).
- (3)
- Proton pump inhibitor (PPI) group. Omeprazole (6.07 mg/kg/d), lansoprazole (4.55 mg/kg/d), esomeprazole (6.07 mg/kg/d), pantoprazole (6.07 mg/kg/d).
- (4)
- Non-steroidal anti-inflammatory (NSAIDs) group. Ibuprofen (91.0 mg/kg/d), diclofenac sodium (15.2 mg/kg/d), naproxen (121.3 mg/kg/d).
2.6. Fluorescence Responses Assay of Detection and Identification of Proteinuria
2.7. Histopathological Analysis
2.8. Statistical Analysis
3. Results
3.1. Characterization of AuNPs–PEI/FLPs Sensor
3.2. Fluorescence Titrations
3.3. Discrimination and Evaluation of the Process of AGs Nephropathy Model
3.4. Discrimination and Evaluation of the Process of PPI Nephropathy Model
3.5. Discrimination and Evaluation of the Process of NSAIDs Nephropathy Model
3.6. Discrimination and Evaluation of the Different Types of DIKI
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, S.Y.; Moon, A. Drug-induced nephrotoxicity and its biomarkers. Biomol. Ther. 2012, 20, 268–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perazella, M.A. Renal vulnerability to drug toxicity. Clin. J. Am. Soc. Nephrol. 2009, 4, 1275–1283. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Huang, J. Drug-Induced Nephrotoxicity: Pathogenic Mechanisms, Biomarkers and Prevention Strategies. Curr. Drug Metab. 2018, 19, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Pavkovic, M.; Vaidya, V.S. MicroRNAs and drug-induced kidney injury. Pharm. Ther. 2016, 163, 48–57. [Google Scholar] [CrossRef] [Green Version]
- Nolin, T.D.; Himmelfarb, J. Mechanisms of drug-induced nephrotoxicity. Handb. Exp. Pharmacol. 2010, 196, 111–130. [Google Scholar]
- Ghane Shahrbaf, F.; Assadi, F. Drug-induced renal disorders. J. Ren. Inj. Prev. 2015, 4, 57–60. [Google Scholar]
- Pazhayattil, G.S.; Shirali, A.C. Drug-induced impairment of renal function. Int. J. Nephrol. Renov. Dis. 2014, 7, 457–468. [Google Scholar]
- Perazella, M.A. Drug-induced nephropathy: An update. Expert. Opin. Drug. Saf. 2005, 4, 689–706. [Google Scholar] [CrossRef]
- Lopez-Novoa, J.M.; Quiros, Y.; Vicente, L.; Morales, A.I.; Lopez-Hernandez, F.J. New insights into the mechanism of aminoglycoside nephrotoxicity: An integrative point of view. Kidney Int. 2011, 79, 33–45. [Google Scholar] [CrossRef] [Green Version]
- Caravaca-Fontán, F.; Fernández-Juárez, G.; Praga, M. Acute kidney injury in interstitial nephritis. Curr. Opin. Crit. Care 2019, 25, 558–564. [Google Scholar] [CrossRef]
- Griffin, B.R.; Faubel, S.; Edelstein, C.L. Biomarkers of Drug-Induced Kidney Toxicity. Ther. Drug Monit. 2019, 41, 213–226. [Google Scholar] [CrossRef] [PubMed]
- Kane-Gill, S.L.; Smithburger, P.L.; Kashani, K.; Kellum, J.A.; Frazee, E. Clinical Relevance and Predictive Value of Damage Biomarkers of Drug-Induced Kidney Injury. Drug Saf. 2017, 40, 1049–1074. [Google Scholar] [CrossRef] [PubMed]
- Redahan, L.; Murray, P.T. Novel Biomarkers of Drug-Induced Kidney Injury. Clin. Pharm. Ther. 2018, 103, 396–398. [Google Scholar] [CrossRef]
- Redahan, L.; Murray, P.T. Biomarkers of drug-induced kidney injury. Curr. Opin. Crit. Care 2017, 23, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.; Miao, Q.; Huang, J.; Li, J.; Pu, K. Multiplex Optical Urinalysis for Early Detection of Drug-Induced Kidney Injury. Anal. Chem. 2020, 92, 6166–6172. [Google Scholar] [CrossRef]
- Haraldsson, B.; Nyström, J.; Deen, W.M. Properties of the glomerular barrier and mechanisms of proteinuria. Physiol. Rev. 2008, 88, 451–487. [Google Scholar] [CrossRef] [Green Version]
- Patrakka, J.; Tryggvason, K. Molecular make-up of the glomerular filtration barrier. Biochem. Biophys. Res. Commun. 2010, 396, 164–169. [Google Scholar] [CrossRef]
- Levey, A.S.; Becker, C.; Inker, L.A. Glomerular filtration rate and albuminuria for detection and staging of acute and chronic kidney disease in adults: A systematic review. JAMA 2015, 313, 837–846. [Google Scholar] [CrossRef] [Green Version]
- Tulkens, P.M. Nephrotoxicity of aminoglycoside antibiotics. Toxicol. Lett. 1989, 46, 107–123. [Google Scholar] [CrossRef]
- Jamshidzadeh, A.; Heidari, R.; Mohammadi-Samani, S.; Azarpira, N.; Najbi, A.; Jahani, P.; Abdoli, N. A comparison between the nephrotoxic profile of gentamicin and gentamicin nanoparticles in mice. J. Biochem. Mol. Toxicol. 2015, 29, 57–62. [Google Scholar] [CrossRef]
- Markowitz, G.S.; Perazella, M.A. Drug-induced renal failure: A focus on tubulointerstitial disease. Clin. Chim. Acta 2005, 351, 31–47. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, D.; Ahmed, Z. Drug-associated renal dysfunction and injury. Nat. Clin. Pract. Nephrol. 2006, 2, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Zhang, Y.; Tang, B.; Zhang, C.Y. Multicolor Quantum Dot-Based Chemical Nose for Rapid and Array-Free Differentiation of Multiple Proteins. Anal. Chem. 2016, 88, 2051–2058. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Goel, H.L.; Le, N.B.; Yoshii, T.; Mout, R.; Tonga, G.Y.; Amante, J.J.; Mercurio, A.M.; Rotello, V.M. Rapid phenotyping of cancer stem cells using multichannel nanosensor arrays. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 1931–1939. [Google Scholar] [CrossRef]
- Pode, Z.; Peri-Naor, R.; Georgeson, J.M.; Ilani, T.; Kiss, V.; Unger, T.; Markus, B.; Barr, H.M.; Motiei, L.; Margulies, D. Protein recognition by a pattern-generating fluorescent molecular probe. Nat. Nanotechnol. 2017, 12, 1161–1168. [Google Scholar] [CrossRef]
- Behera, P.; De, M. Nano-Graphene Oxide Based Multichannel Sensor Arrays towards Sensing of Protein Mixtures. Chem. Asian J. 2019, 14, 553–560. [Google Scholar] [CrossRef]
- Li, X.; Kong, H.; Mout, R.; Saha, K.; Moyano, D.F.; Robinson, S.M.; Rana, S.; Zhang, X.; Riley, M.A.; Rotello, V.M. Rapid identification of bacterial biofilms and biofilm wound models using a multichannel nanosensor. ACS Nano 2014, 8, 12014–12019. [Google Scholar] [CrossRef] [Green Version]
- Rana, S.; Elci, S.G.; Mout, R.; Singla, A.K.; Yazdani, M.; Bender, M.; Bajaj, A.; Saha, K.; Bunz, U.H.F.; Jirik, F.R.; et al. Ratiometric Array of Conjugated Polymers-Fluorescent Protein Provides a Robust Mammalian Cell Sensor. J. Am. Chem. Soc. 2016, 138, 4522–4529. [Google Scholar] [CrossRef] [Green Version]
- Le, N.D.B.; Yesilbag Tonga, G.; Mout, R.; Kim, S.T.; Wille, M.E.; Rana, S.; Dunphy, K.A.; Jerry, D.J.; Yazdani, M.; Ramanathan, R.; et al. Cancer Cell Discrimination Using Host-Guest “Doubled” Arrays. J. Am. Chem. Soc. 2017, 139, 8008–8012. [Google Scholar] [CrossRef]
- Xu, S.; Gao, T.; Feng, X.; Fan, X.; Liu, G.; Mao, Y.; Yu, X.; Lin, J.; Luo, X. Near infrared fluorescent dual ligand functionalized Au NCs based multidimensional sensor array for pattern recognition of multiple proteins and serum discrimination. Biosens. Bioelectron. 2017, 97, 203–207. [Google Scholar] [CrossRef]
- Yuan, Z.; Du, Y.; Tseng, Y.T.; Peng, M.; Cai, N.; He, Y.; Chang, H.-T.; Yeung, E.S. Fluorescent gold nanodots based sensor array for proteins discrimination. Anal. Chem. 2015, 87, 4253–4259. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lu, Y. Preparation of aptamer-linked gold nanoparticle purple aggregates for colorimetric sensing of analytes. Nat. Protoc. 2006, 1, 246–252. [Google Scholar] [CrossRef] [PubMed]










Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, K.; Wang, B.; Lin, J.; Han, L.; Li, M.; Wang, P.; Yu, X.; Tian, J. A Multichannel Fluorescent Array Sensor for Discrimination of Different Types of Drug-Induced Kidney Injury. Sensors 2023, 23, 6114. https://doi.org/10.3390/s23136114
Sun K, Wang B, Lin J, Han L, Li M, Wang P, Yu X, Tian J. A Multichannel Fluorescent Array Sensor for Discrimination of Different Types of Drug-Induced Kidney Injury. Sensors. 2023; 23(13):6114. https://doi.org/10.3390/s23136114
Chicago/Turabian StyleSun, Kunhui, Bing Wang, Jiaoli Lin, Lei Han, Meifang Li, Ping Wang, Xiean Yu, and Jiangwei Tian. 2023. "A Multichannel Fluorescent Array Sensor for Discrimination of Different Types of Drug-Induced Kidney Injury" Sensors 23, no. 13: 6114. https://doi.org/10.3390/s23136114
APA StyleSun, K., Wang, B., Lin, J., Han, L., Li, M., Wang, P., Yu, X., & Tian, J. (2023). A Multichannel Fluorescent Array Sensor for Discrimination of Different Types of Drug-Induced Kidney Injury. Sensors, 23(13), 6114. https://doi.org/10.3390/s23136114