Algorithms for Liver Segmentation in Computed Tomography Scans: A Historical Perspective
Abstract
:1. Introduction
- RQ1—What are the challenges and limitations associated with accurate liver segmentation in CT scans?
- RQ2—How does the choice of the method impact the accuracy and efficiency of liver segmentation in CT scans?
- RQ3—What are the evaluation metrics commonly used to assess the performance of AI models and traditional methods for liver segmentation in CT scans?
2. Methodology
2.1. Data Sources
2.2. Search Queries
2.3. Inclusion Criteria
2.4. Exclusion Criteria
2.5. Characterisation of Selected Papers
3. Literature Review
3.1. Historical Overview
3.2. Other Review Papers
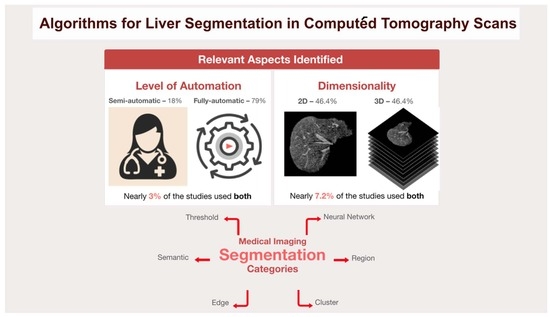
4. Findings
5. Discussion
5.1. Public Dataset Analysis
5.2. Impact of the Adoption of Neuronal-Network-Based Methods
5.3. Comparison between 2D and 3D Methods for Liver Segmentation
- Importance of Choosing between 2D and 3D Methods
- -
- In medical imaging, and in particular liver segmentation, the choice between slice-based 2D and volume-based 3D segmentation methods is crucial. This decision is highly dependent on the anatomical structure of the liver. Given the complex, three-dimensional nature of the liver, 3D segmentation techniques often prove to be the most appropriate choice [21,40]. These methods are inherently designed to understand and process the volumetric characteristics of the liver, which is a critical consideration for accurate segmentation results.
- Two-Dimensional Segmentation Limitations
- -
- Although 2D slice-based segmentation is widely used, it has limitations, particularly when it comes to dealing with complex organs such as the liver. The main challenge with 2D methods is their inability to fully capture all the regions of the liver. They involve working with individual slices, which can provide a fragmented understanding of the organ structure, but this fragmentation can lead to inconsistencies and errors when these individual slices are aggregated to form a complete image [41].
- Three-Dimensional Segmentation Advantages
- -
- In order to overcome the limitations of 2D segmentation, 3D segmentation has the ability to use more contextual information. Unlike 2D methods, which visualise the liver in individual slices, 3D techniques consider the organ as a whole, as they have the ability to ensure anatomical correctness by processing the liver as a single, continuous volume, avoiding errors that can arise from the aggregation of 2D slices [14,39]. In 2D segmentation, inconsistencies can occur when individual slices are combined, leading to inaccuracies in the representation of the liver anatomy. The holistic view provided by 3D segmentation results in more accurate segmentation, as it takes into account the spatial relationships and continuity between the different sections of the liver. The inclusion of this additional contextual information can potentially lead to segmentation results, especially in complex cases where the shape and size of the liver can vary considerably.
5.4. Exploring Research Questions
- RQ1—What are the challenges and limitations associated with accurate liver segmentation in CT scans?
- -
- The challenges and limitations associated with accurate liver segmentation in CT images include under-segmentation, over-segmentation, low contrast, poor boundary detection, and background segmentation due to noise. In addition, liver segmentation in CT scans is further challenged by the presence of artefacts, such as partial volumes, noise, and low sharpness and contrast between organs, making it difficult to identify the boundaries between different tissues.
- RQ2—How does the choice of the method impact the accuracy and efficiency of liver segmentation in CT scans?
- -
- The choice of the method has an important impact on the accuracy and efficiency of liver segmentation in CT scans. Traditional techniques such as image processing and region growing approaches have shown varying degrees of sensitivity and specificity, with some challenges in dealing with large injuries. In contrast, newer methods such as FCN and DBN-DNN and techniques like ResU-Net and SegNet showed a higher accuracy, with some reaching the highest accuracy levels. Notably, the use of GPUs has reduced processing times, thus contributing towards more efficient and accurate liver segmentation methods.
- RQ3—What are the evaluation metrics commonly used to assess the performance of AI models and traditional methods for liver segmentation in CT scans?
- -
- Some of the key metrics used to measure the outcome of segmentation techniques include the Dice Similarity Coefficient (DSC), accuracy, precision, sensitivity, specificity, and the segmentation speed. There is not much consistency in the metrics presented by the various studies except for the DSC.
6. Conclusions and Future Work
Author Contributions
Funding
Conflicts of Interest
References
- Francque, S. The Liver and the Cardiovascular System: Two of a Kind? J. Am. Heart Assoc. 2021, 10, e020286. [Google Scholar] [CrossRef]
- Trefts, E.; Gannon, M.; Wasserman, D.H. The liver. Curr. Biol. 2017, 27, R1147–R1151. [Google Scholar] [CrossRef]
- Mendes, B.; Domingues, I.; Silva, A.; Santos, J. Prostate Cancer Aggressiveness Prediction Using CT Images. Life 2021, 11, 1164. [Google Scholar] [CrossRef]
- Pereira, G.; Domingues, I.; Martins, P.; Abreu, P.H.; Duarte, H.; Santos, J. Registration of CT with PET: A Comparison of Intensity-Based Approaches. In International Workshop on Combinatorial Image Analysis (IWCIA); Springer: Berlin/Heidelberg, Germany, 2018; pp. 134–149. [Google Scholar]
- Zhang, H.; Luo, K.; Deng, R.; Li, S.; Duan, S. Deep Learning-Based CT Imaging for the Diagnosis of Liver Tumor. Comput. Intell. Neurosci. 2022, 2022, 1–7. [Google Scholar] [CrossRef]
- Vernuccio, F.; Cannella, R.; Bartolotta, T.V.; Galia, M.; Tang, A.; Brancatelli, G. Advances in liver US, CT, and MRI: Moving toward the future. Eur. Radiol. Exp. 2021, 5, 52. [Google Scholar] [CrossRef] [PubMed]
- Domingues, I.; Cardoso, J.S. Using Bayesian surprise to detect calcifications in mammogram images. In Proceedings of the 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Chicago, IL, USA, 26–30 August 2014; pp. 1091–1094. [Google Scholar]
- Mendes, B.; Domingues, I.; Santos, J. Multi-class Semantic Segmentation for Prostate Cancer Radiotherapy Treatment Optimization. In Proceedings of the International Conference on Mathematical Analysis and Applications in Science and Engineering (ICMA2SC), Porto, Portugal, 27–29 June 2022. [Google Scholar]
- Bechar, M.E.A.; Settouti, N.; Domingues, I. Deep Learning vs. Super Pixel Classification for Breast Masses Segmentation. In Deep Learning for Biomedical Applications; CRC Press: Boca Raton, FL, USA, 2021; pp. 121–156. [Google Scholar]
- Oliveira, A.C.; Domingues, I.; Duarte, H.; Santos, J.; Abreu, P.H. Going Back to Basics on Volumetric Segmentation of the Lungs in CT: A Fully Image Processing Based Technique. In Iberian Conference on Pattern Recognition and Image Analysis (IbPRIA); Springer: Berlin/Heidelberg, Germany, 2019; Volume 11868 LNCS, pp. 322–334. [Google Scholar]
- Carbone, I.; Martins, P.; Teixeira, A.; Silva, A. A Vocal Tract Segmentation and Analysis over a European Portuguese MRI Database. Electrónica E Telecomunicações 2008, 4, 1050–1053. [Google Scholar]
- Zhou, L.Q.; Wang, J.Y.; Yu, S.Y.; Wu, G.G.; Wei, Q.; Deng, Y.B.; Wu, X.L.; Cui, X.W.; Dietrich, C.F. Artificial intelligence in medical imaging of the liver. World J. Gastroenterol. 2019, 25, 672–682. [Google Scholar] [CrossRef] [PubMed]
- Chlebus, G.; Schenk, A.; Moltz, J.H.; van Ginneken, B.; Hahn, H.K.; Meine, H. Automatic liver tumor segmentation in CT with fully convolutional neural networks and object-based postprocessing. Sci. Rep. 2018, 8, 15497. [Google Scholar] [CrossRef] [PubMed]
- Christ, P.F.; Ettlinger, F.; Grün, F.; Elshaera, M.E.A.; Lipkova, J.; Schlecht, S.; Ahmaddy, F.; Tatavarty, S.; Bickel, M.; Bilic, P.; et al. Automatic Liver and Tumor Segmentation of CT and MRI Volumes using Cascaded Fully Convolutional Neural Networks. arXiv 2017, arXiv:1702.05970. [Google Scholar] [CrossRef]
- Ansari, M.Y.; Abdalla, A.; Ansari, M.Y.; Ansari, M.I.; Malluhi, B.; Mohanty, S.; Mishra, S.; Singh, S.S.; Abinahed, J.; Al-Ansari, A.; et al. Practical utility of liver segmentation methods in clinical surgeries and interventions. BMC Med. Imaging 2022, 22, 97. [Google Scholar]
- Le, D.C.; Chinnasarn, K.; Chansangrat, J.; Keeratibharat, N.; Horkaew, P. Semi-automatic liver segmentation based on probabilistic models and anatomical constraints. Sci. Rep. 2021, 11, 6106. [Google Scholar] [CrossRef] [PubMed]
- Halevi, G.; Moed, H.; Bar-Ilan, J. Suitability of Google Scholar as a source of scientific information and as a source of data for scientific evaluation—Review of the Literature. J. Inf. 2017, 11, 823–834. [Google Scholar] [CrossRef]
- Sakshi; Kukreja, V. Image Segmentation Techniques: Statistical, Comprehensive, Semi-Automated Analysis and an Application Perspective Analysis of Mathematical Expressions. Arch. Comput. Methods Eng. 2023, 30, 457–495. [Google Scholar] [CrossRef]
- Bae, K.T.; Giger, M.L.; Chen, C.T.; Kahn, C.E., Jr. Automatic segmentation of liver structure in CT images. Med. Phys. 1993, 20, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Heath, D.G.; Kuszyk, B.S.; Fishman, E.K. Automatic liver segmentation technique for three-dimensional visualization of CT data. Radiology 1996, 201, 359–364. [Google Scholar] [CrossRef]
- Soler, L.; Malandain, G.; Montagnat, J.; Delingette, H.; Ayache, N.; Clément, J.M.; Roy, C.; Russier, Y.; Tassetti, V.; Marescaux, J. Automatic Segmentation of Portal Vein in CT-Scans of the Liver. In Proceedings of the World Congress on Medical Physics and Biomedical Engineering (MPBE), Nice, France, 14–19 September 1997; p. 788. [Google Scholar]
- Yoo, S.W.; Cho, J.S.; Noh, S.M.; Shin, K.S.; Park, J.W. Advanced Liver Segmentation by Using Pixel Ratio in Abdominal CT Image. In Proceedings of the IEEK Conference, Pusan, Korea, 2000; The Institute of Electronics and Information Engineers: Seoul, Korea, 2000; pp. 39–42. [Google Scholar]
- Pan, S.; Dawant, B.M. Automatic 3D segmentation of the liver from abdominal CT images: A level-set approach. In Medical Imaging: Image Processing; SPIE: Bellingham, WA, USA, 2001; Volume 4322, pp. 128–138. [Google Scholar]
- Saitoh, T.; Tamura, Y.; Kaneko, T. Automatic segmentation of liver region through blood vessels on multi-phase CT. In Proceedings of the International Conference on Pattern Recognition, Quebec, ON, Canada, 11–15 August 2002; IEEE: Piscataway, NJ, USA, 2002; Volume 1, pp. 735–738. [Google Scholar]
- Masumoto, J.; Hori, M.; Sato, Y.; Murakami, T.; Johkoh, T.; Nakamura, H.; Tamura, S. Automated liver segmentation using multislice CT images. Syst. Comput. Jpn. 2003, 34, 71–82. [Google Scholar] [CrossRef]
- Lim, S.J.; Jeong, Y.Y.; Lee, C.W.; Ho, Y.S. Automatic segmentation of the liver in CT images using the watershed algorithm based on morphological filtering. In Proceedings of the Medical Imaging: Image Processing, San Diego, CA, USA, 6–19 February 2004; SPIE: Bellingham, WA, USA, 2004; Volume 5370, pp. 1658–1666. [Google Scholar]
- Liu, F.; Zhao, B.; Kijewski, P.K.; Wang, L.; Schwartz, L.H. Liver segmentation for CT images using GVF snake. Med. Phys. 2005, 32, 3699–3706. [Google Scholar] [CrossRef]
- Lim, S.J.; Jeong, Y.Y.; Ho, Y.S. Automatic liver segmentation for volume measurement in CT Images. J. Vis. Commun. Image Represent. 2006, 17, 860–875. [Google Scholar] [CrossRef]
- Beichel, R.; Bauer, C.; Bornik, A.; Sorantin, E.; Bischof, H. Liver Segmentation in CT Data: A Segmentation Refinement Approach. In Segmentation in The Clinic: A Grand Challenge; 2007; pp. 235–245. Available online: https://graz.elsevierpure.com/en/publications/liver-segmentation-in-ct-data-a-segmentation-refinement-approach (accessed on 4 March 2024).
- Massoptier, L.; Casciaro, S. A new fully automatic and robust algorithm for fast segmentation of liver tissue and tumors from CT scans. Eur. Radiol. 2008, 18, 1658–1665. [Google Scholar] [CrossRef]
- Heimann, T.; Van Ginneken, B.; Styner, M.A.; Arzhaeva, Y.; Aurich, V.; Bauer, C.; Beck, A.; Becker, C.; Beichel, R.; Bekes, G.; et al. Comparison and evaluation of methods for liver segmentation from CT datasets. IEEE Trans. Med Imaging 2009, 28, 1251–1265. [Google Scholar] [CrossRef]
- Akram, M.U.; Khanum, A.; Iqbal, K. An automated system for liver CT enhancement and segmentation. Icgst J. Graph. Vis. Image Process. (ICGST-GVIP) 2010, 10, 17–22. [Google Scholar]
- Oliveira, D.A.; Feitosa, R.Q.; Correia, M.M. Segmentation of liver, its vessels and lesions from CT images for surgical planning. Biomed. Eng. Online 2011, 10, 1–23. [Google Scholar] [CrossRef]
- Linguraru, M.G.; Richbourg, W.J.; Liu, J.; Watt, J.M.; Pamulapati, V.; Wang, S.; Summers, R.M. Tumor Burden Analysis on Computed Tomography by Automated Liver and Tumor Segmentation. IEEE Trans. Med. Imaging 2012, 31, 1965–1976. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Luo, S.; Li, J. Liver Segmentation from CT Image Using Fuzzy Clustering and Level Set. J. Signal Inf. Process. 2013, 4, 36–42. [Google Scholar] [CrossRef]
- Jena, B.; Krishna Nayak, G.; Saxena, S. Comprehensive Review of Abdominal Image Segmentation using Soft and Hard Computing Approaches. In Proceedings of the International Conference on Computer Science, Engineering and Applications (ICCSEA), Gunupur, India, 13–14 March 2020; pp. 1–5. [Google Scholar]
- Platero, C.; Tobar, M.C. A multiatlas segmentation using graph cuts with applications to liver segmentation in CT scans. Comput. Math. Methods Med. 2014, 2014, 182909. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, A.; Fouad, A.; Abd Elfattah, M.; Hassanien, A.E.; Hefny, H.; Zhu, S.Y.; Schaefer, G. CT liver segmentation using artificial bee colony optimisation. Procedia Comput. Sci. 2015, 60, 1622–1630. [Google Scholar] [CrossRef]
- Dou, Q.; Chen, H.; Jin, Y.; Yu, L.; Qin, J.; Heng, P.A. 3D deeply supervised network for automatic liver segmentation from CT volumes. In Proceedings of the 19th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Athens, Greece, 17–21 October 2016; Springer: Berlin/Heidelberg, Germany, 2016; pp. 149–157. [Google Scholar]
- Hiraman, A. Liver Segmentation Using 3D CT Scans. Ph.D. Thesis, University of Kwazulu-Natal, Durban, South Africa, 2018. [Google Scholar]
- Wang, K.; Mamidipalli, A.; Retson, T.; Bahrami, N.; Hasenstab, K.; Blansit, K.; Bass, E.; Delgado, T.; Cunha, G.; Middleton, M.S.; et al. Automated CT and MRI liver segmentation and biometry using a generalized convolutional neural network. Radiol. Artif. Intell. 2019, 1, 180022. [Google Scholar] [CrossRef]
- Almotairi, S.; Kareem, G.; Aouf, M.; Almutairi, B.; Salem, M.A.M. Liver tumor segmentation in CT scans using modified SegNet. Sensors 2020, 20, 1516. [Google Scholar] [CrossRef]
- Soler, L.; Hostettler, A.; Agnus, V.; Charnoz, A.; Fasquel, J.; Moreau, J.; Osswald, A.; Bouhadjar, M.; Marescaux, J. 3D Image Reconstruction for Comparison of Algorithm Database: A Patient Specific Anatomical and Medical Image Database; Technical Report; IRCAD: Strasbourg, France, 2010. [Google Scholar]
- Ayalew, Y.A.; Fante, K.A.; Mohammed, M.A. Modified U-Net for liver cancer segmentation from computed tomography images with a new class balancing method. BMC Biomed. Eng. 2021, 3, 4. [Google Scholar] [CrossRef]
- LiTS Challenge Dataset. Available online: https://competitions.codalab.org/competitions/17094 (accessed on 27 November 2023).
- Scicluna, D. Automatic Segmentation of Healthy Liver in Abdominal Computed Tomography Scans. Master’s Thesis, University of Malta, Msida, Malta, 2022. [Google Scholar]
- Kavur, A.E.; Gezer, N.S.; Barış, M.; Aslan, S.; Conze, P.H.; Groza, V.; Pham, D.D.; Chatterjee, S.; Ernst, P.; Özkan, S.; et al. CHAOS Challenge-combined (CT-MR) healthy abdominal organ segmentation. Med. Image Anal. 2021, 69, 101950. [Google Scholar] [CrossRef]
- Ezzat, K.A.; Omran, L.N.; El Seddawy, A.I.B. Automatic liver segmentation in computed tomography scans using deep semantic segmentation. Bull. Electr. Eng. Inform. 2023, 12, 250–256. [Google Scholar] [CrossRef]
- Shao, J.; Luan, S.; Ding, Y.; Xue, X.; Zhu, B.; Wei, W. Attention Connect Network for Liver Tumor Segmentation from CT and MRI Images. Technol. Cancer Res. Treat. 2024, 23, 15330338231219366. [Google Scholar] [PubMed]
- Maurício, J.; Domingues, I.; Bernardino, J. Comparing Vision Transformers and Convolutional Neural Networks for Image Classification: A Literature Review. Appl. Sci. 2023, 13, 5521. [Google Scholar] [CrossRef]
- Priyadarsini, S.; Selvathi, D. Survey on segmentation of liver from CT images. In Proceedings of the IEEE International Conference on Advanced Communication Control and Computing Technologies (ICACCCT), Ramanathapuram, India, 23–25 August 2012; pp. 234–238. [Google Scholar]
- Mharib, A.M.; Ramli, A.R.; Mashohor, S.; Mahmood, R.B. Survey on liver CT image segmentation methods. Artif. Intell. Rev. 2012, 37, 83–95. [Google Scholar] [CrossRef]
- Khan, F. Automated segmentation of CT liver images: A review. J. Commun. Technol. Electron. Comput. Sci. 2018, 19, 5–9. [Google Scholar]
- Moghbel, M.; Mashohor, S.; Mahmud, R.; Saripan, M.I.B. Review of liver segmentation and computer assisted detection/diagnosis methods in computed tomography. Artif. Intell. Rev. 2018, 50, 497–537. [Google Scholar] [CrossRef]
- Vanmore, S.V.; Chougule, S.R. Survey on automatic liver segmentation techniques from abdominal CT images. In Proceedings of the International Conference on Intelligent Computing and Control Systems (ICCS), Madurai, India, 22–24 May 2024; IEEE: Piscataway, NJ, USA, 2019; pp. 1030–1035. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25. [Google Scholar] [CrossRef]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. (IJCV) 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015, arXiv:1505.04597. [Google Scholar] [CrossRef]
- Mir, R.; Kelly, S.M.; Xiao, Y.; Moore, A.; Clark, C.H.; Clementel, E.; Corning, C.; Ebert, M.; Hoskin, P.; Hurkmans, C.W.; et al. Organ at risk delineation for radiation therapy clinical trials: Global Harmonization Group consensus guidelines. Radiother. Oncol. J. Eur. Soc. Ther. Radiol. Oncol. 2020, 150, 30–39. [Google Scholar]
- Sarria, G.R.; Kugel, F.; Roehner, F.; Layer, J.; Dejonckheere, C.; Scafa, D.; Koeksal, M.; Leitzen, C.; Schmeel, L.C. Artificial Intelligence–Based Autosegmentation: Advantages in Delineation, Absorbed Dose-Distribution, and Logistics. Adv. Radiat. Oncol. 2024, 9, 101394. [Google Scholar] [PubMed]
- Al-Saeed, Y.; Gab-Allah, W.; Elmogy, M. Hepatic tumors diagnosis system based on fuzzy c-means using computed tomography images. Res. Sq. Prepr. 2022. [Google Scholar] [CrossRef]
- MICCAI-Sliver07. Available online: https://sliver07.grand-challenge.org/ (accessed on 27 November 2023).








| Authors | Year | Segmentation Category | Method | Autom. Level | Dim. | Database | Results |
|---|---|---|---|---|---|---|---|
| Bae et al. [19] | 1993 | Threshold | Grey-level thresholding | Semi | 2D | Private | 0.985 DSC with mean percent error within 10%. |
| Gao et al. [20] | 1996 | Edge | Parametrically deformable contour model | Fully | 3D | Private | 13.2% of the results required user modifications. |
| Soler et al. [21] | 1997 | Edge | Deformable models | Fully | 3D | Private | Claimed to be comparable to manual segmentations. |
| Yoo et al. [22] | 2000 | Threshold | Threshold | Fully | 2D | Private | 3.41% error. |
| Pan and Dawant [23] | 2001 | Edge | Level sets | Fully | Both | Private | [0.874, 0.963] average similarities. |
| Saitoh et al. [24] | 2002 | Threshold | Threshold | Fully | 3D | Private | ∼20 min computation time. |
| Masumoto et al. [25] | 2003 | Region | Differences between time-phase images | Fully | 3D | Private | 67% volume ratio average; 32% in the worst cases. |
| Lim et al. [26] | 2004 | Region | Watershed | Fully | 2D | Private | Only qualitative. |
| Liu et al. [27] | 2005 | Edge | GVF snake | Semi | 2D | Private | 5.3% median value of the difference ratios. |
| Lim et al. [28] | 2006 | Semantic | Labeling-based search | Fully | 2D | Private | 96% average correctness; 3% average error rate. |
| Beichel et al. [29] | 2007 | Region | Graph cuts | Semi | 3D | Private | 5.2% average overlap error. |
| Massoptier and Casciaro [30] | 2008 | Edge | Active contour | Fully | 3D | Private | 94.2% mean DSC. |
| Heimann et al. [31] | 2009 | Several | Majority Voting | Both | Both | Private | 5% overlap error; −0.7 volume difference; 0.8 average distance; 1.7 RMS distance; 19.1 max distance. |
| Akram et al. [32] | 2010 | Threshold | Global Threshold | Fully | 3D | Private | 0.96 average accuracy; 0.0017 std; 96% accurately segmented; 4% poorly segmented. |
| Oliveira et al. [33] | 2011 | Edge | Level sets | Semi | 2D | SLiver07 | 82.05 overall score. |
| Linguraru et al. [34] | 2012 | Region | Graph cuts | Fully | 3D | Private; SLiver07 | 2.2 VOE. |
| Li et al. [35] | 2013 | Edge | Fuzzy clustering and level set | Fully | 2D | Private | 0.9986 average accuracy; 0.9989 average specificity. |
| Platero et al. [37] | 2014 | Region | Graph cuts | Semi | 3D | SLiver07 | 76.3 maximum score; 0.973 DSC. |
| Mostafa et al. [38] | 2015 | Cluster | ABC optimization | Fully | 2D | Private | 93.73% accuracy; 84.82% average SI. |
| Dou et al. [39] | 2016 | NN | 3D DSN | Fully | 3D | SLiver07 | 5.42% VOE; 0.79 mm ASSD. |
| Christ et al. [14] | 2017 | NN | CFCN | Fully | 2D | 3D-IRCADb01 | 94.3% mean DSC. |
| Hiraman [40] | 2018 | NN | CNN | Fully | 2D | SLiver07 | 12.07% average VOE; −1.96% RVD; 2.25 mm ASSD; 2.60 mm RMSD; 43.01 mm MSSD. |
| Wang et al. [41] | 2019 | NN | CNN | Fully | 3D | Private | DSC. |
| Almotairi et al. [42] | 2020 | NN | SegNet | Fully | 3D | 3D-IRCADb01 | 94.57% overall accuracy. |
| Ayalew et al. [44] | 2021 | NN | U-Net | Fully | 2D | 3D-IRCADb01; LiTS | 0.9612 DSC. |
| Scicluna [46] | 2022 | NN | UNet; VGG16UNetC | Fully | 2D | CHAOS | 85.84 mean score; 97.85 DSC; 80.33 RAVD; 94.80 ASSD score; 70.38 MSSD. |
| Ezzat et al. [48] | 2023 | NN | CNN | Fully | 2D | Private | 98.80% accuracy. |
| Shao et al. [49] | 2024 | NN | AC-Net | Fully | 3D | Private; LiTS | 0.90 DSC; 0.82 JC; 0.92 recall; 0.89 precision; 11.96 HD; 4.59 ASSD. |
| Category | Description | Main Advantages | Main Limitations | Applicability |
|---|---|---|---|---|
| Threshold | Segments based on intensity thresholds | Simple, fast, easy to implement | Sensitivity to threshold selection, suffers from noise and artefacts | Commonly used in cases where clear intensity differences between the ROI and other regions exist |
| Edge | Segments based on intensity transitions between regions | Accurate delineation of organ boundaries and structures | Sensitive to noise, difficulties with capturing complex structures | Suitable for images with clear organ boundaries and well-defined edges, but may struggle with low-contrast areas |
| Region | Segments based on homogeneous regions within an image | Simple implementation, intuitive methodology | Sensitive to initialisation | Often used in cases where interpretability is a concern, but may struggle with fine details |
| Semantic | Segments based on semantic meaning of pixels | Pixel-level segmentation, fine-grained structural detail | Complex to implement, resource-intensive, and computationally expensive | Suitable for segmenting anatomical structures with distinct features |
| Cluster | Segments based on similar data patterns or clusters | Efficient grouping and identification of similar data patterns | Sensitivity to initialisation and noise, limited to specific data distributions | Useful for identifying patterns and groups within the data, but can struggle with irregular shapes |
| NN | Learns models to segment images based on learned features | High accuracy, efficient learning from data | Requires large training datasets, computationally intensive | Suitable for different types of data due to its flexibility and adaptability |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niño, S.B.; Bernardino, J.; Domingues, I. Algorithms for Liver Segmentation in Computed Tomography Scans: A Historical Perspective. Sensors 2024, 24, 1752. https://doi.org/10.3390/s24061752
Niño SB, Bernardino J, Domingues I. Algorithms for Liver Segmentation in Computed Tomography Scans: A Historical Perspective. Sensors. 2024; 24(6):1752. https://doi.org/10.3390/s24061752
Chicago/Turabian StyleNiño, Stephanie Batista, Jorge Bernardino, and Inês Domingues. 2024. "Algorithms for Liver Segmentation in Computed Tomography Scans: A Historical Perspective" Sensors 24, no. 6: 1752. https://doi.org/10.3390/s24061752
APA StyleNiño, S. B., Bernardino, J., & Domingues, I. (2024). Algorithms for Liver Segmentation in Computed Tomography Scans: A Historical Perspective. Sensors, 24(6), 1752. https://doi.org/10.3390/s24061752