Bioassay-Guided Isolation of 2-[p-(2-Carboxyhydrazino)phenoxy]-6-(hydroxymethyl)tetrahydro-2H-pyran-3,4,5-triol from Oroxylum indicum and the Investigation of Its Molecular Mechanism Action of Apoptosis Induction
Abstract
:1. Introduction
2. Results
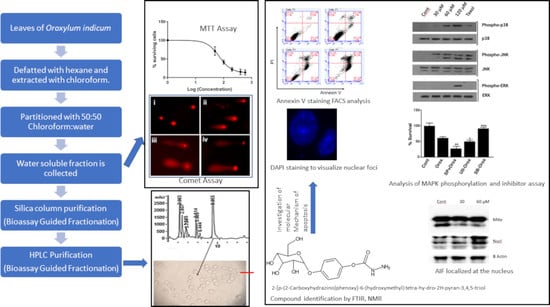
2.1. Leaf Crude Extract of OI Inhibits the Proliferation of PC3 Cells
2.2. The Leaf Crude Extract of OI Induces Apoptosis in PC3 Cells
2.3. Bioassay-Guided Fractionation Purified a Novel Bioactive Compound
2.4. Chemical Characterization
2.5. Oroxyquinone Induces Apoptosis in PC3 Cells
2.6. Oroxyquinone Induces Cell Cycle Arrest in PC3 Cells
2.7. Oroxyquinone Inhibits PC3 Cell Migration
2.8. Oroxyquinone Induces Cells Apoptosis in PC3 Cells in a Caspase-3-Independent Mechanism
2.9. Oroxyquinone Activates MAPK Proteins
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Extraction and Purification
4.3. Cell Viability Assay (MTT Assay)
4.4. gDNA Fragmentation Assay
4.5. Chemical Characterization
4.6. Comet Assay
4.7. Annexin-V Analysis
4.8. Wound Healing Assay
4.9. Cellular Fractionation
4.10. Western Blots
4.11. Cell CYCLE Analysis
4.12. DAPI Staining
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sener, S.F.; Grey, N. The global burden of cancer. J. Surg. Oncol. 2005, 92, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Vitale, I.; Abrams, J.; Alnemri, E.; Baehrecke, E.; Blagosklonny, M.; Dawson, T.M.; Dawson, V.; El-Deiry, W.; Fulda, S.J.C.D.; et al. Molecular definitions of cell death subroutines: Recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012, 19, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Harminder; Singh, V.; Chaudhary, A.K. A Review on the Taxonomy, Ethnobotany, Chemistry and Pharmacology of Oroxylum indicum Vent. Indian J. Pharm. Sci. 2011, 73, 483–490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa-Lotufo, L.V.; Khan, M.T.; Ather, A.; Wilke, D.V.; Jimenez, P.C.; Pessoa, C.; de Moraes, M.E.; de Moraes, M.O. Studies of the anticancer potential of plants used in Bangladeshi folk medicine. J. Ethnopharmacol. 2005, 99, 21–30. [Google Scholar] [CrossRef]
- Moirangthem, D.S.; Talukdar, N.C.; Bora, U.; Kasoju, N.; Das, R.K. Differential effects of Oroxylum indicum bark extracts: Antioxidant, antimicrobial, cytotoxic and apoptotic study. Cytotechnology 2013, 65, 83–95. [Google Scholar] [CrossRef] [Green Version]
- Nakahara, K.; Onishi-Kameyama, M.; Ono, H.; Yoshida, M.; Trakoontivakorn, G. Antimutagenic activity against trp-P-1 of the edible Thai plant, Oroxylum indicum vent. Biosci. Biotechnol. Biochem. 2001, 65, 2358–2360. [Google Scholar] [CrossRef]
- Naveen Kumar, D.R.; Cijo George, V.; Suresh, P.K.; Ashok Kumar, R. Cytotoxicity, apoptosis induction and anti-metastatic potential of Oroxylum indicum in human breast cancer cells. Asian Pac. J. Cancer Prev. APJCP 2012, 13, 2729–2734. [Google Scholar] [CrossRef] [Green Version]
- Li, N.-n.; Meng, X.-s.; Men, W.-x.; Bao, Y.-r.; Wang, S.J.E.-B.C.; Medicine, A. Total flavonoids from Oroxylum indicum induce apoptosis via PI3K/akt/PTEN signaling pathway in liver cancer. Evid.-Based Complement. Altern. Med. 2018, 2018. [Google Scholar] [CrossRef] [Green Version]
- Lalrinzuali, K.; Vabeiryureilai, M.; Jagetia, G.C.J.B.C. Sonapatha (Oroxylum indicum) mediates cytotoxicity in cultured HeLa cells by inducing apoptosis and suppressing NF-κB, COX-2, RASSF7 and NRF2. Bioorganic Chem. 2021, 114, 105126. [Google Scholar] [CrossRef]
- Kruger, A.; Ganzera, M. Oroxylum indicum seeds—Analysis of flavonoids by HPLC-MS. J. Pharm. Biomed. Anal. 2012, 70, 553–556. [Google Scholar] [CrossRef]
- Wahab, N.H.; Mat, N.F.C. Baicalein-rich fraction of Oroxylum indicum leaves induces apoptosis by repressing E6 and E7 expression in HPV-associated cervical cancer cell lines. Int. J. Res. Pharm. Sci. 2018, 9, 108–117. [Google Scholar]
- Wahab, N.H.; Mat, N.F.C. Baicalein-rich fraction of Oroxylum indicum triggers mitochondrial-mediated apoptosis pathway through MAPK transduction in cervical cancer cells. Malays. J. Fundam. Appl. Sci. 2021, 17, 39–43. [Google Scholar] [CrossRef]
- Buranrat, B.; Noiwetch, S.; Suksar, T.; Ta-Ut, A. Inhibition of cell proliferation and migration by Oroxylum indicum extracts on breast cancer cells via Rac1 modulation. J. Pharm. Anal. 2020, 10, 187–193. [Google Scholar] [CrossRef]
- Jabbar, S.; Khan, M.T.; Choudhuri, M.S.; Sil, B.K. Bioactivity studies of the individual ingredients of the Dashamularishta. Pak. J. Pharm. Sci. 2004, 17, 9–17. [Google Scholar]
- Sharma, J.; Gairola, S.; Gaur, R.D.; Painuli, R.M.; Siddiqi, T.O. Ethnomedicinal plants used for treating epilepsy by indigenous communities of sub-Himalayan region of Uttarakhand, India. J. Ethnopharmacol. 2013, 150, 353–370. [Google Scholar] [CrossRef]
- Tran, T.V.; Malainer, C.; Schwaiger, S.; Hung, T.; Atanasov, A.G.; Heiss, E.H.; Dirsch, V.M.; Stuppner, H. Screening of Vietnamese medicinal plants for NF-kappaB signaling inhibitors: Assessing the activity of flavonoids from the stem bark of Oroxylum indicum. J. Ethnopharmacol. 2015, 159, 36–42. [Google Scholar] [CrossRef] [Green Version]
- Siriwatanametanon, N.; Fiebich, B.L.; Efferth, T.; Prieto, J.M.; Heinrich, M. Traditionally used Thai medicinal plants: In vitro anti-inflammatory, anticancer and antioxidant activities. J. Ethnopharmacol. 2010, 130, 196–207. [Google Scholar] [CrossRef]
- Mat Ali, R.; Houghton, P.J.; Raman, A.; Hoult, J.R. Antimicrobial and antiinflammatory activities of extracts and constituents of Oroxylum indicum (L.) Vent. Phytomed. Int. J. Phytother. Phytopharm. 1998, 5, 375–381. [Google Scholar] [CrossRef]
- Mao, A. Oroxylum indicum vent—A potential anticancer medicinal plant. Ind. J. Trad. Knowl. 2002, 1, 17–21. [Google Scholar]
- Gu, Q.; Luo, H.; Zheng, W.; Liu, Z.; Huang, Y. Pseudonocardia oroxyli sp. nov., a novel actinomycete isolated from surface-sterilized Oroxylum indicum root. Int. J. Syst. Evol. Microbiol. 2006, 56, 2193–2197. [Google Scholar] [CrossRef] [Green Version]
- Joshi, S.V.; Vyas, B.A.; Shah, P.D.; Shah, D.R.; Shah, S.A.; Gandhi, T.R. Protective effect of aqueous extract of Oroxylum indicum Linn. (root bark) against DNBS-induced colitis in rats. Indian J. Pharmacol. 2011, 43, 656–661. [Google Scholar] [CrossRef]
- Radhika, L.G.; Meena, C.V.; Peter, S.; Rajesh, K.S.; Rosamma, M.P. Phytochemical and antimicrobial study of Oroxylum indicum. Anc. Sci. Life 2011, 30, 114–120. [Google Scholar]
- Doshi, K.; Ilanchezhian, R.; Acharya, R.; Patel, B.R.; Ravishankar, B. Anti-inflammatory activity of root bark and stem bark of Shyonaka. J. Ayurveda Integr. Med. 2012, 3, 194–197. [Google Scholar] [CrossRef] [Green Version]
- Sharma, J.; Gairola, S.; Gaur, R.D.; Painuli, R.M. The treatment of jaundice with medicinal plants in indigenous communities of the Sub-Himalayan region of Uttarakhand, India. J. Ethnopharmacol. 2012, 143, 262–291. [Google Scholar] [CrossRef]
- Karnati, M.; Chandra, R.H.; Veeresham, C.; Kishan, B. Anti-arthritic activity of root bark of Oroxylum indicum (L.) vent against adjuvant-induced arthritis. Pharmacogn. Res. 2013, 5, 121–128. [Google Scholar] [CrossRef] [Green Version]
- Rajkumar, V.; Guha, G.; Kumar, R.A. Isolation and bioactivity evaluation of two metabolites from the methanolic extract of Oroxylum indicum stem bark. Asian Pac. J. Trop. Biomed. 2012, 2, S7–S11. [Google Scholar] [CrossRef]
- Lalou, C.; Basak, A.; Mishra, P.; Mohanta, B.C.; Banik, R.; Dinda, B.; Khatib, A.M. Inhibition of tumor cells proliferation and migration by the flavonoid furin inhibitor isolated from Oroxylum indicum. Curr. Med. Chem. 2013, 20, 583–591. [Google Scholar]
- Singh, L.S.; Berk, M.; Oates, R.; Zhao, Z.; Tan, H.; Jiang, Y.; Zhou, A.; Kirmani, K.; Steinmetz, R.; Lindner, D.; et al. Ovarian cancer G protein-coupled receptor 1, a new metastasis suppressor gene in prostate cancer. J. Natl. Cancer Inst. 2007, 99, 1313–1327. [Google Scholar] [CrossRef]
- Mathiasen, I.S.; Jäättelä, M. Triggering caspase-independent cell death to combat cancer. Trends Mol. Med. 2002, 8, 212–220. [Google Scholar] [CrossRef]
- Tait, S.W.; Green, D.R. Caspase-independent cell death: Leaving the set without the final cut. Oncogene 2008, 27, 6452–6461. [Google Scholar] [CrossRef] [Green Version]
- Tait, S.W.; Green, D.R. Mitochondrial regulation of cell death. Cold Spring Harb. Perspect. Biol. 2013, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishra, A.P.; Salehi, B.; Sharifi-Rad, M.; Pezzani, R.; Kobarfard, F.; Sharifi-Rad, J.; Nigam, M. Programmed cell death, from a cancer perspective: An overview. Mol. Diagn. Ther. 2018, 22, 281–295. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.R.; Fu, Y.S.; Tsai, M.J.; Cheng, H.; Weng, C.F. Natural compounds from herbs that can potentially execute as autophagy inducers for cancer therapy. Int. J. Mol. Sci. 2017, 18, 1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddivari, L.; Vanamala, J.; Chintharlapalli, S.; Safe, S.H.; Miller, J.C. Anthocyanin fraction from potato extracts is cytotoxic to prostate cancer cells through activation of caspase-dependent and caspase-independent pathways. Carcinogenesis 2007, 28, 2227–2235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Liu, H.; Tian, Z.; Griffith, B.N.; Ji, M.; Li, Q.Q. Gossypol induces apoptosis in human PC-3 prostate cancer cells by modulating caspase-dependent and caspase-independent cell death pathways. Life Sci. 2007, 80, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Hilchie, A.L.; Furlong, S.J.; Sutton, K.; Richardson, A.; Robichaud, M.R.; Giacomantonio, C.A.; Ridgway, N.D.; Hoskin, D.W. Curcumin-induced apoptosis in PC3 prostate carcinoma cells is caspase-independent and involves cellular ceramide accumulation and damage to mitochondria. Nutr. Cancer 2010, 62, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Debroy, S.; Kramarenko, I.I.; Ghose, S.; Oleinik, N.V.; Krupenko, S.A.; Krupenko, N.I. A novel tumor suppressor function of glycine N-methyltransferase is independent of its catalytic activity but requires nuclear localization. PLoS ONE 2013, 8, e70062. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Jiang, Y.; Li, Z.; Kravchenko, V.V.; Ulevitch, R.J. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature 1997, 386, 296–299. [Google Scholar] [CrossRef]
- Wada, T.; Penninger, J.M. Mitogen-activated protein kinases in apoptosis regulation. Oncogene 2004, 23, 2838–2849. [Google Scholar] [CrossRef] [Green Version]
- Werlen, G.; Hausmann, B.; Naeher, D.; Palmer, E.J.S. Signaling life and death in the thymus: Timing is everything. Science 2003, 299, 1859–1863. [Google Scholar] [CrossRef]
- Rudolf, E.; Kralova, V.; Rudolf, K.; John, S. The role of p38 in irinotecan-induced DNA damage and apoptosis of colon cancer cells. Mutat. Res. 2013, 741–742, 27–34. [Google Scholar] [CrossRef]
- Rudolf, E.; Rudolf, K.; Červinka, M. Selenium activates p53 and p38 pathways and induces caspase-independent cell death in cervical cancer cells. Cell Biol. Toxicol. 2008, 24, 123–141. [Google Scholar] [CrossRef]
- Chen, W.; Liu, L.; Luo, Y.; Odaka, Y.; Awate, S.; Zhou, H.; Shen, T.; Zheng, S.; Lu, Y.; Huang, S. Cryptotanshinone activates p38/JNK and inhibits Erk1/2 leading to caspase-independent cell death in tumor cells. Cancer Prev. Res. 2012, 5, 778–787. [Google Scholar] [CrossRef] [Green Version]
- Kamalden, T.A.; Ji, D.; Osborne, N.N. Rotenone-induced death of RGC-5 cells is caspase independent, involves the JNK and p38 pathways and is attenuated by specific green tea flavonoids. Neurochem. Res. 2012, 37, 1091–1101. [Google Scholar] [CrossRef]
- Kwon, C.H.; Yoon, C.S.; Kim, Y.K.J.T. Ciglitazone induces caspase-independent apoptosis via p38-dependent AIF nuclear translocation in renal epithelial cells. Neurochem. Res. 2008, 244, 13–24. [Google Scholar] [CrossRef]
- Park, D.; Patek, P. Detergent and enzyme treatment of apoptotic cells for the observation of DNA fragmentation. Biotechniques 1998, 24, 558. [Google Scholar] [CrossRef]
- Olive, P.L.; Banáth, J.P. The comet assay: A method to measure DNA damage in individual cells. Nat. Protoc. 2006, 1, 23. [Google Scholar] [CrossRef]
- Rodriguez, L.G.; Wu, X.; Guan, J.-L. Wound-healing assay. Cell Migr. 2005, 294, 23–29. [Google Scholar]











Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, A.R.; Singh, S.A.; Singh, T.D.; Singh, N.T.; Machathoibi, T.C.; Singh, O.M.; Singh, L.S. Bioassay-Guided Isolation of 2-[p-(2-Carboxyhydrazino)phenoxy]-6-(hydroxymethyl)tetrahydro-2H-pyran-3,4,5-triol from Oroxylum indicum and the Investigation of Its Molecular Mechanism Action of Apoptosis Induction. Pharmaceuticals 2022, 15, 559. https://doi.org/10.3390/ph15050559
Singh AR, Singh SA, Singh TD, Singh NT, Machathoibi TC, Singh OM, Singh LS. Bioassay-Guided Isolation of 2-[p-(2-Carboxyhydrazino)phenoxy]-6-(hydroxymethyl)tetrahydro-2H-pyran-3,4,5-triol from Oroxylum indicum and the Investigation of Its Molecular Mechanism Action of Apoptosis Induction. Pharmaceuticals. 2022; 15(5):559. https://doi.org/10.3390/ph15050559
Chicago/Turabian StyleSingh, Asem Robinson, Salam Asbin Singh, Thangjam Davis Singh, Naorem Tarundas Singh, Takhellambam Chanu Machathoibi, Okram Mukherjee Singh, and Lisam Shanjukumar Singh. 2022. "Bioassay-Guided Isolation of 2-[p-(2-Carboxyhydrazino)phenoxy]-6-(hydroxymethyl)tetrahydro-2H-pyran-3,4,5-triol from Oroxylum indicum and the Investigation of Its Molecular Mechanism Action of Apoptosis Induction" Pharmaceuticals 15, no. 5: 559. https://doi.org/10.3390/ph15050559
APA StyleSingh, A. R., Singh, S. A., Singh, T. D., Singh, N. T., Machathoibi, T. C., Singh, O. M., & Singh, L. S. (2022). Bioassay-Guided Isolation of 2-[p-(2-Carboxyhydrazino)phenoxy]-6-(hydroxymethyl)tetrahydro-2H-pyran-3,4,5-triol from Oroxylum indicum and the Investigation of Its Molecular Mechanism Action of Apoptosis Induction. Pharmaceuticals, 15(5), 559. https://doi.org/10.3390/ph15050559