Activation of the Silent Secondary Metabolite Production by Introducing Neomycin-Resistance in a Marine-Derived Penicillium purpurogenum G59
Abstract
:1. Introduction

2. Results and Discussion
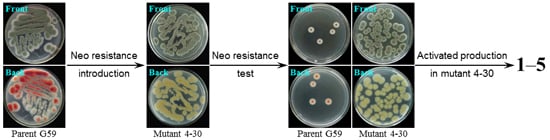
2.1. Preliminary Test and Mutant Selection
| Group | DMSO% (v/v) | Neomycin (mg/mL) | DMSO% Tested on G59 Strain | Treatment Times (Day) |
|---|---|---|---|---|
| I | 20% | 2.0 | Previously in [13] | 1–60 |
| II | 33% | 3.3 | Newly in present study | 1–60 |
| III | 50% | 5.0 | Previously in [13] | 1–60 |
| IV | 67% | 6.7 | Newly in present study | 1–60 |
| V | 100% | 10.0 | Newly in present study | 1–60 |

| Group | DMSO% (v/v) | Neomycin (mg/mL) | Treatment Time at 4 °C (Day) | Total | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 Day | 2 Day | 5 Day | 7 Day | 10 Day | 15 Day | 30 Day | 60 Day | ||||
| I | 20% | 2.0 | NS | NS | NS | NS | NS | 1 | 1 | 1 | 3 |
| II | 33% | 3.3 | NS | NS | NS | 1 | 2 | 2 | 1 | 2 | 8 |
| III | 50% | 5.0 | NS | 2 | 2 | 3 | 4 | 4 | 2 | 1 | 18 |
| IV | 67% | 6.7 | NS | 1 | 3 | 4 | 4 | 2 | NC | NC | 14 |
| V | 100% | 10.0 | 2 | 1 | 3 | 5 | 2 | NC | NC | NC | 13 |
| Total | 2 | 4 | 8 | 13 | 12 | 9 | 4 | 4 | 56 | ||

2.2. Resistance Test




2.3. Estimation of Activated Bioactive Metabolite Production in Mutants by Bioassay
| Strain | Condition for Treatment of the G59 Spores at 4 °C to Select the Mutant | Inhibition Rate (IR%) at 100 μg/mL | |||||
|---|---|---|---|---|---|---|---|
| DMSO% (v/v) | Neomycin (mg/mL) | Treatment Time (Day) | First | Second | Third | Mean ± SD | |
| G59 | – | – | – | 4.8 | 3.9 | 7.9 | 5.5 ± 2.1 |
| 1-50-1 | 20 | 2 | 15 | 39.8 | 35.3 | 45.5 | 40.2 ± 5.1 |
| 2-2 | 33 | 3.3 | 10 | 27.7 | 47.6 | 30.9 | 35.4 ± 10.7 |
| 2-2-3 | 33 | 3.3 | 10 | 27.0 | 34.5 | 34.6 | 32.0 ± 4.4 |
| 2-50-1 | 33 | 3.3 | 15 | 32.6 | 31.4 | 19.8 | 27.9 ± 7.1 |
| 2-s-2 | 33 | 3.3 | 60 | 47.7 | 73.2 | 38.3 | 53.1 ± 18.1 |
| 2-s-3 | 33 | 3.3 | 60 | 46.0 | 59.2 | 36.0 | 47.1 ± 11.6 |
| 3-4-1 | 50 | 5.0 | 5 | 34.5 | 35.2 | 44.7 | 38.1 ± 5.7 |
| 3-4-2 | 50 | 5.0 | 5 | 30.8 | 31.5 | 41.8 | 34.7 ± 6.2 |
| 3-50-1 | 50 | 5.0 | 10 | 43.5 | 27.7 | 53.1 | 41.4 ± 12.8 |
| 3-50-2 | 50 | 5.0 | 10 | 69.2 | 31.7 | 40.6 | 47.2 ± 19.6 |
| 3-f-1 | 50 | 5.0 | 15 | 24.8 | 53.4 | 40.3 | 39.5 ± 14.3 |
| 3-f-3 | 50 | 5.0 | 15 | 31.6 | 33.8 | 31.5 | 35.2 ± 1.3 |
| 3-f-31 | 50 | 5.0 | 15 | 55.1 | 29.9 | 38.5 | 41.2 ± 12.8 |
| 3-v-1 | 50 | 5.0 | 30 | 30.4 | 35.3 | 44.9 | 36.9 ± 7.4 |
| 3-x-1 | 50 | 5.0 | 30 | 40.3 | 70.1 | 49.7 | 53.4 ± 15.2 |
| 3-s-1 | 50 | 5.0 | 60 | 24.0 | 24.7 | 34.5 | 27.7 ± 5.9 |
| 3-s-2 | 50 | 5.0 | 60 | 55.1 | 23.1 | 49.8 | 42.7 ± 17.2 |
| 4-4 | 67 | 6.7 | 5 | 42.5 | 34.3 | 30.7 | 35.8 ± 6.0 |
| 4-16-2 | 67 | 6.7 | 7 | 29.4 | 32.8 | 29.1 | 30.4 ± 2.1 |
| 4-30 | 67 | 6.7 | 7 | 50.4 | 53.1 | 36.1 | 46.5 ± 9.1 |
| 4-x-1 | 67 | 6.7 | 10 | 47.3 | 34.9 | 51.8 | 44.7 ± 8.8 |
| 4-v-2 | 67 | 6.7 | 10 | 34.9 | 38.6 | 38.9 | 37.5 ± 2.2 |
| 4-s-1 | 67 | 6.7 | 15 | 68.1 | 51.4 | 27.5 | 49.0 ± 20.4 |
| PDN-10-2 | 100 | 10 | 1 | 41.3 | 52.4 | 23.8 | 39.2 ± 14.4 |
| PDN-50-1 | 100 | 10 | 5 | 52.8 | 25.8 | 47.2 | 41.9 ± 14.2 |
| PDN-v-1 | 100 | 10 | 7 | 29.3 | 22.9 | 26.0 | 26.1 ± 3.2 |
| PDN-v-2 | 100 | 10 | 7 | 51.2 | 21.8 | 36.1 | 36.4 ± 14.7 |
| PDN-s-2 | 100 | 10 | 10 | 38.8 | 35.4 | 32.2 | 35.5 ± 3.3 |
2.4. Chromatographic Analysis of the Activated Secondary Metabolite Production in the Mutants

2.5. Bioactive Metabolites 1–5 Newly Produced by Introducing Drug-Resistance in Mutant 4-30
2.5.1. Fermentation, Isolation and Identification
2.5.2. Inhibitory Effect of 1–5 on Several Human Cancer Cell Lines
| Compound | K562 | HL-60 | HeLa | BGC-823 |
|---|---|---|---|---|
| 1 | 73.7% | 70.0% | 68.8% | 66.0% |
| 2 | 60.1% | 79.4% | 51.9% | 67.1% |
| 3 | 77.5% | 82.0% | 84.1% | 79.9% |
| 4 | 33.1% | 43.3% | 38.0% | 27.5% |
| 5 | 87.5% | 86.5% | 88.5% | 86.4% |
| Compound | K562 | HL-60 | HeLa | BGC-823 |
|---|---|---|---|---|
| 1 | 80.1 (274.3) | 85.2 (291.8) | 88.1 (301.7) | 85.1 (291.4) |
| 2 | 58.2 (232.8) | 44.4 (177.6) | 58.3 (233.2) | 57.1 (228.4) |
| 3 | 50.8 (133.7) | 43.2 (113.7) | 65.6 (172.6) | 54.2 (142.6) |
| 5 | 27.2 (61.5) | 15.4 (34.8) | 20.8 (47.1) | 24.1 (54.5) |
2.5.3. HPLC-PDAD-UV/HPLC-ESI-MS Analyses for Detecting 1–5 in the Mutant 4-30 Extract
2.6. Discussion
3. Experimental Section
3.1. General Experimental
3.2. MTT Assay
3.3. Experiments for Introducing Neomycin Resistance into G59 Strain to Activate Silent Metabolites
3.3.1. Initial Strain and Spore Suspension Preparation
3.3.2. Treatment of G59 Spores with Neomycin Coupled with DMSO and Mutant Selection
| Group | Concentration of DMSO and Neo in Test Group | Mixed Volume (mL) of Solution or Solvent with G59 Spore Suspension for I–IV Groups | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Test Group | Neo Control | DMSO Control | Blank Control | |||||||
| DMSO% | Neo mg/mL | Neo-D | G59-S | Neo-H | G59-S | DMSO | G59-S | H2O | G59-S | |
| I | 20% | 2.0 | 0.6 | 2.4 | 0.6 | 2.4 | 0.6 | 2.4 | 0.6 | 2.4 |
| II | 33% | 3.3 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| III | 50% | 5.0 | 1.5 | 1.5 | 1.5 | 1.5 | 1.5 | 1.5 | 1.5 | 1.5 |
| IV | 67% | 6.7 | 2.0 | 1.0 | 2.0 | 1.0 | 2.0 | 1.0 | 2.0 | 1.0 |
| V | 100% | 10.0 | Approx. eq. amt. of G59 spores in each 3.0 mL of Neo-D, Neo-H, DMSO and H2O. | |||||||
3.3.3. Resistance Test for Acquired Resistance of Four Mutants to Neomycin
3.3.4. Fermentation and Preparation of EtOAc Extract for MTT Assay and Chemical Analysis
3.3.5. HPLC-PDAD-UV and HPLC-ESI-MS Analyses
3.4. Experiments for Investigation on Compounds 1–5 from Mutant 4-30
3.4.1. Large-Scale Fermentation and EtOAc Extract Preparation
3.4.2. Isolation of Compounds 1–5
3.4.3. Physicochemical and Spectroscopic Data of 1–5
3.4.4. Examination of 1–5 in EtOAc Extracts of Mutant 4-30 and Parent G59 Strain
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pejin, B.; Jovanović, K.K.; Mojović, M.; Savić, A.G. New and highly potent antitumor natural products from marine-derived fungi: Covering the period from 2003 to 2012. Curr. Top. Med. Chem. 2013, 13, 2745–2766. [Google Scholar] [CrossRef] [PubMed]
- Rateb, M.E.; Ebel, R. Secondary metabolites of fungi from marine habitats. Nat. Prod. Rep. 2011, 28, 290–344. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, I.; Kim, S.-K. Immense essence of excellence: Marine microbial bioactive compounds. Mar. Drugs 2010, 8, 2673–2701. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Marine-sourced anti-cancer and cancer pain control agents in clinical and late preclinical development. Mar. Drugs 2014, 12, 255–278. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2014, 31, 160–258. [Google Scholar] [CrossRef] [PubMed]
- Hertweck, C. Hidden biosynthetic treasures brought to light. Nat. Chem. Biol. 2009, 5, 450–452. [Google Scholar] [CrossRef] [PubMed]
- Brakhage, A.A.; Schroeckh, V. Fungal secondary metabolites—Strategies to activate silent gene clusters. Fungal Genet. Biol. 2011, 48, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Cichewicz, R.H.; Henrikson, J.C.; Wang, X.; Branscum, K.M. Strategies for accessing microbial secondary metabolites from silent biosynthetic pathways. In Manual of Industrial Microbiology and Biotechnology, 3rd ed.; Baltz, R.H., Davies, J.E., Demain, A.L., Bull, A.T., Junker, B., Katz, L., Lynd, L.R., Masurekar, P.C., Reeves, D., Zhao, H., Eds.; ASM Press: Washington, DC, USA, 2010; pp. 78–95. [Google Scholar]
- Chiang, Y.-M.; Chang, S.-L; Oakley, B.R.; Wang, C.C.C. Recent advances in awakening silent biosynthetic gene clusters and linking orphan clusters to natural products in microorganisms. Curr. Opin. Chem. Biol. 2011, 15, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Ochi, K.; Hosaka, T. New strategies for drug discovery: Activation of silent or weakly expressed microbial gene clusters. Appl. Microbiol. Biotechnol. 2013, 97, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Marmann, A.; Aly, A.H.; Lin, W.; Wang, B.; Proksch, P. Co-cultivation—A powerful emerging tool for enhancing the chemical diversity of microorganisms. Mar. Drugs 2014, 12, 1043–1065. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, J.A.; Teles, A.P.C.; Bracarense, A.A.P.; Gomes, D.C. Classical and epigenetic approaches to metabolite diversification in filamentous fungi. Phytochem. Rev. 2013. [Google Scholar] [CrossRef]
- Chai, Y.-J.; Cui, C.-B.; Li, C.-W.; Wu, C.-J.; Tian, C.-K.; Hua, W. Activation of the dormant secondary metabolite production by introducing gentamicin-resistance in a marine-derived Penicillium purpurogenum G59. Mar. Drugs 2012, 10, 559–582. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.-M.; Wu, C.-J.; Li, C.-W.; Cui, C.-B. A practical strategy to discover new antitumor compounds by activating silent metabolite production in fungi by diethyl sulphate mutagenesis. Mar. Drugs 2014, 12, 1788–1814. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Cui, C.-B.; Li, C.-W.; Hua, W.; Wu, C.-J.; Zhu, T.-J.; Gu, Q.-Q. Activation of dormant secondary metabolite production by introducing neomycin resistance into the deep-sea fungus, Aspergillus versicolor ZBY-3. Mar. Drugs 2014, 12, 4326–4352. [Google Scholar] [CrossRef] [PubMed]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Henrikson, J.C.; Hoover, A.R.; Joyner, P.M.; Cichewicz, R.H. A chemical epigenetics approach for engineering the in situbiosynthesis of a cryptic natural product from Aspergillus niger. Org. Biomol. Chem. 2009, 7, 435–438. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.-M.; Cui, C.-B.; Li, C.-W.; Wu, C.-J.; Zhang, Z.-J.; Li, L.; Huang, X.-J.; Ye, W.-C. Purpurogemutantin and purpurogemutantidin, new drimenyl cyclohexenone derivatives produced by a mutant obtained by diethyl sulfate mutagenesis of a marine-derived Penicillium purpurogenum G59. Mar. Drugs 2012, 10, 1266–1287. [Google Scholar] [CrossRef] [PubMed]
- Xia, M.-W.; Cui, C.-B.; Li, C.-W.; Wu, C.-J. Three new and eleven known unusual C25 steroids: Activated production of silent metabolites in a marine-derived fungus by chemical mutagenesis strategy using diethyl sulphate. Mar. Drugs 2014, 12, 1545–1568. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-J.; Li, C.-W.; Cui, C.-B. Seven new and two known lipopeptides as well as five known polyketides: The activated production of silent metabolites in a marine-derived fungus by chemical mutagenesis strategy using diethyl sulphate. Mar. Drugs 2014, 12, 1815–1838. [Google Scholar] [CrossRef] [PubMed]
- Ochi, K.; Okamoto, S.; Tozawa, Y.; Inaoka, T.; Hosaka, T.; Xu, J.; Kurosawa, K. Ribosome engineering and secondary metabolite production. Adv. Appl. Microbiol. 2004, 56, 155–184. [Google Scholar] [PubMed]
- Ochi, K. From microbial differentiation to ribosome engineering. Biosci. Biothenol. Biochem. 2007, 71, 1373–1386. [Google Scholar] [CrossRef]
- Hosaka, T.; Ohnishi-Kameyama, M.; Muramatsu, H.; Murakami, K.; Tsurumi, Y.; Kodani, S.; Yoshida, M.; Fujie, A.; Ochi, K. Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat. Biotechnol. 2009, 27, 462–464. [Google Scholar] [CrossRef] [PubMed]
- Fu, P.; Jamison, M.; La, S.; MacMillan, J.B. Inducamides A–C, chlorinated alkaloids from an RNA polymerase mutant strain of Streptomyces sp. Org. Lett. 2014, 16, 5656–5659. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.K.; Cui, C.B.; Han, X.X. Isolation of fungal strains in unusual environment and screening for their antitumor activity. J. Int. Pharm. Res. 2008, 35, 401–405. [Google Scholar]
- Chai, Y.J.; Cui, C.B.; Li, C.W.; Hua, W. Antitumor metabolites newly produced by a gentamicin-resistant mutant of Penicillium purpurogenum G59. J. Int. Pharm. Res. 2011, 38, 216–222. [Google Scholar]
- Chen, X.-W.; Li, C.-W.; Cui, C.-B.; Hua, W.; Zhu, T.-J.; Gu, Q.-Q. Nine new and five known polyketides derived from a deep sea-sourced Aspergillus sp. 16-02-1. Mar. Drugs 2014, 12, 3116–3137. [Google Scholar] [CrossRef] [PubMed]
- Ghisalberti, E.L.; Hockless, D.C.R.; Rowland, C.Y.; White, A.H. Structural study of curvularin, a cell division inhibitor. Aust. J. Chem. 1993, 46, 571–575. [Google Scholar]
- Wakana, D.; Hosoe, T.; Itabashi, T.; Okada, K.; Takaki, G.M.C.; Yaguchi, T.; Fukushima, K.; Kawai, K. New citrinin derivatives isolated from Penicillium citrinum. J. Nat. Med. 2006, 60, 279–284. [Google Scholar] [CrossRef]
- Gao, H.; Hong, K.; Chen, G.D.; Wang, C.X.; Tang, J.S.; Yu, Y.; Jiang, M.M.; Li, M.M.; Wang, N.L.; Yao, X.S. New oxidized sterols from Aspergillus awamori and the endo-boat conformation adopted by the cyclohexene oxide system. Magn. Reson. Chem. 2010, 48, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Espinosa-Artiles, P.; Schubert, V; Xu, Y.-M.; Zhang, W.; Lin, M.; Gunatilaka, A.A.L.; Süssmuth, R.; Molnár, I. Characterization of the biosynthetic genes for 10,11-dehydrocurvularin, a heat-shock response modulator anticancer fungal polyketide from Aspergillus terreus. Appl. Environ. Microbiol. 2013, 79, 2038–2047. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Kinoshita, H.; Ishihara, S.; Sakai, K.; Nagai, S.; Nihira, T. Polyketide synthase gene responsible for citrinin biosynthesis in Monascus purpureus. Appl. Environ. Microbiol. 2005, 71, 3453–3457. [Google Scholar] [CrossRef] [PubMed]
- Marinhoa, A.M.R.; Rodrigues-Filho, E.; Ferreira, A.G.; Santos, L.S. C25 steroid epimers produced by Penicillium janthinellum, a fungus isolated from fruits Melia azedarach. J. Braz. Chem. Soc. 2005, 16, 1342–1346. [Google Scholar] [CrossRef]
- Lees, N.D.; Skaggs, B.; Kitsch, D.R.; Bard, M. Cloning of the late genes in the ergosterol biosynthetic pathway of Saccharomyces cerevisiae—A review. Lipids 1995, 30, 221–226. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, C.-J.; Yi, L.; Cui, C.-B.; Li, C.-W.; Wang, N.; Han, X. Activation of the Silent Secondary Metabolite Production by Introducing Neomycin-Resistance in a Marine-Derived Penicillium purpurogenum G59. Mar. Drugs 2015, 13, 2465-2487. https://doi.org/10.3390/md13042465
Wu C-J, Yi L, Cui C-B, Li C-W, Wang N, Han X. Activation of the Silent Secondary Metabolite Production by Introducing Neomycin-Resistance in a Marine-Derived Penicillium purpurogenum G59. Marine Drugs. 2015; 13(4):2465-2487. https://doi.org/10.3390/md13042465
Chicago/Turabian StyleWu, Chang-Jing, Le Yi, Cheng-Bin Cui, Chang-Wei Li, Nan Wang, and Xiao Han. 2015. "Activation of the Silent Secondary Metabolite Production by Introducing Neomycin-Resistance in a Marine-Derived Penicillium purpurogenum G59" Marine Drugs 13, no. 4: 2465-2487. https://doi.org/10.3390/md13042465
APA StyleWu, C. -J., Yi, L., Cui, C. -B., Li, C. -W., Wang, N., & Han, X. (2015). Activation of the Silent Secondary Metabolite Production by Introducing Neomycin-Resistance in a Marine-Derived Penicillium purpurogenum G59. Marine Drugs, 13(4), 2465-2487. https://doi.org/10.3390/md13042465