Enhanced Lipid Production and Molecular Dynamics under Salinity Stress in Green Microalga Chlamydomonas reinhardtii (137C)
Abstract
:1. Introduction
2. Results and Discussion
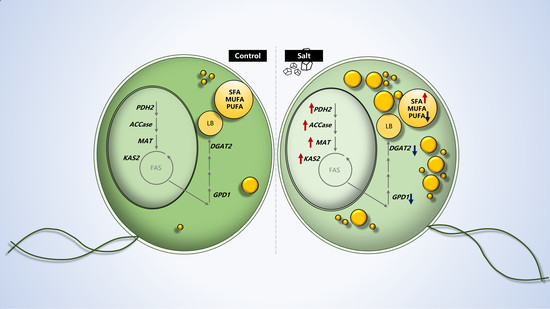
2.1. Salts Trigger Intracellular Lipid Accumulation in C. reinhardtii (137c)
2.2. Dynamics of Intracellular Fatty Acids during Salt Stress
2.3. Gene Expression Analysis
2.4. Generation of Expressing Cyanobacterium Cells Carrying ChMAT and SynMAT
2.5. MAT Transcription was Up-Regulated Depending on Growth Phase in Expressing Cells
2.6. Lipid Analysis in Transformants
3. Materials and Methods
3.1. Strains and Growth Conditions
3.2. Stress Treatments
3.3. Lipid Analysis
3.4. Gene Expression Analysis
3.5. Construction of Expressing Cells
3.6. Other Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lang, I.; Hodac, L.; Friedl, T.; Feussner, I. Fatty acid profiles and their distribution patterns in microalgae: A comprehensive analysis of more than 2000 strains from the SAG culture collection. BMC Plant Biol. 2011, 11, 124. [Google Scholar] [CrossRef] [PubMed]
- Martins, D.; Custódio, L.; Barreira, L.; Pereira, H.; Ben-Hamadou, R.; Varela, J.; Abu-Salah, K. Alternative sources of n-3 long-chain polyunsaturated fatty acids in marine microalgae. Mar. Drugs 2013, 11, 2259–2281. [Google Scholar] [CrossRef] [PubMed]
- Ndimba, B.K.; Ndimba, R.J.; Johnson, T.S.; Waditee-Sirisattha, R.; Baba, M.; Sirisattha, S.; Shiraiwa, Y.; Agrawal, G.K.; Rakwal, R. Biofuels as a sustainable energy source: An update of the applications of proteomics in bioenergy crops and algae. J. Proteomics 2013, 93, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Mata, T.M.; Martins, A.A.; Caetano, N.S. Microalgae for biodiesel production and other applications: A review. Renew. Sustain. Energy Rev. 2010, 14, 217–232. [Google Scholar] [CrossRef] [Green Version]
- Noraini, M.; Ong, H.C.; Badrul, M.J.; Chong, W. A review on potential enzymatic reaction for biofuel production from algae. Renew. Sustain. Energy Rev. 2014, 39, 24–34. [Google Scholar] [CrossRef]
- Sakamoto, K.; Baba, M.; Suzuki, I.; Watanabe, M.M.; Shiraiwa, Y. Optimization of light for growth, photosynthesis, and hydrocarbon production by the colonial microalga Botryococcus braunii BOT-22. Bioresour. Technol. 2012, 110, 474–479. [Google Scholar] [CrossRef] [PubMed]
- Zalutskaya, Z.; Kharatyan, N.; Forchhammer, K.; Ermilova, E. Reduction of PII signaling protein enhances lipid body production in Chlamydomonas reinhardtii. Plant Sci. 2015, 240, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.P.; Slabas, A.R.; Rafferty, J.B. Fatty acid biosynthesis in plants—Metabolic pathways, structure and organization. In Lipids in Photosynthesis; Springer: Dordrecht, The Netherlands, 2009; pp. 11–34. [Google Scholar]
- Rastogi, R.P.; Pandey, A.; Larroche, C.; Madamwar, D. Algal Green Energy–R&D and technological perspectives for biodiesel production. Renew. Sustain. Energy Rev. 2018, 82, 2946–2969. [Google Scholar] [CrossRef]
- Zienkiewicz, K.; Du, Z.Y.; Ma, W.; Vollheyde, K.; Benning, C. Stress-induced neutral lipid biosynthesis in microalgae—Molecular, cellular and physiological insights. BBA Mol. Cell Biol. Lipids 2016, 1861, 1269–1281. [Google Scholar] [CrossRef]
- Ho, S.H.; Nakanishi, A.; Ye, X.; Chang, J.S.; Hara, K.; Hasunuma, T.; Kondo, A. Optimizing biodiesel production in marine Chlamydomonas sp. JSC4 through metabolic profiling and an innovative salinity-gradient strategy. Biotechnol. Biofuels 2014, 7, 97. [Google Scholar] [CrossRef]
- Ho, S.H.; Nakanishi, A.; Kato, Y.; Yamasaki, H.; Chang, J.S.; Misawa, N.; Hirose, Y.; Minagawa, J.; Hasunuma, T.; Kondo, A. Dynamic metabolic profiling together with transcription analysis reveals salinity-induced starch-to-lipid biosynthesis in alga Chlamydomonas sp. JSC4. Sci. Rep. 2017, 7, 45471. [Google Scholar] [CrossRef] [PubMed]
- Campenni, L.; Nobre, B.P.; Santos, C.A.; Oliveira, A.; Aires-Barros, M.; Palavra, A.; Gouveia, L. Carotenoid and lipid production by the autotrophic microalga Chlorella protothecoides under nutritional, salinity, and luminosity stress conditions. Appl. Microbiol. Biotechnol. 2013, 97, 1383–1393. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, A.; Aikawa, S.; Ho, S.H.; Chen, C.Y.; Chang, J.S.; Hasunuma, T.; Kondo, A. Development of lipid productivities under different CO2 conditions of marine microalgae Chlamydomonas sp. JSC4. Bioresour. Technol. 2014, 152, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.H.; Jiang, J.G. Advancing oleaginous microorganisms to produce lipid via metabolic engineering technology. Prog. Lipid Res. 2013, 52, 395–408. [Google Scholar] [CrossRef] [PubMed]
- Dawut, K.; Sirisattha, S.; Hibino, T.; Kageyama, H.; Waditee-Sirisattha, R. Functional characterization of the NhaA Na+/H+ antiporter from the green picoalga Ostreococcus tauri. Arch. Biochem. Biophys. 2018, 649, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Harris, E.H. The Chlamydomonas Sourcebook: Introduction to Chlamydomonas and Its Laboratory Use, 2nd ed.; Academic Press: New York, NY, USA, 2009. [Google Scholar]
- Wei, S.; Bian, Y.; Zhao, Q.; Chen, S.; Mao, J.; Song, C.; Cheng, K.; Xiao, Z.; Zhang, C.; Ma, W. Salinity-induced palmella formation mechanism in halotolerant algae Dunaliella salina revealed by quantitative proteomics and phosphoproteomics. Front. Plant Sci. 2017, 8, 810. [Google Scholar] [CrossRef] [PubMed]
- Herron, M.D.; Borin, J.M.; Boswell, J.C.; Walker, J.; Chen, I.C.K.; Knox, C.A.; Boyd, M.; Rosenzweig, F.; Ratcliff, W.C. De novo origins of multicellularity in response to predation. Sci. Rep. 2019, 9, 2328. [Google Scholar] [CrossRef]
- Lurling, M.; Beekman, W. Palmelloids formation in Chlamydomonas reinhardtii: Defence against rotifer predators? Ann. Limnol. Int. J. Limnol. 2006, 42, 65–72. [Google Scholar] [CrossRef]
- Santos, J.M.; Freire, P.; Vicente, M.; Arraiano, C.M. The stationary-phase morphogene bolA from Escherichia coli is induced by stress during early stages of growth. Mol. Microbiol. 1999, 32, 789–798. [Google Scholar] [CrossRef]
- Khona, D.K.; Dongre, S.S.; Arraiano, C.M.; D’Souza, J.S. A BolA-like morphogene from the alga Chlamydomonas reinhardtii changes morphology and induces biofilm formation in Escherichia coli. FEMS Microbiol. Lett. 2013, 339, 39–47. [Google Scholar] [CrossRef]
- Sato, N.; Tsuzuki, M.; Matsuda, Y.; Ehara, T.; Osafune, T.; Kawaguchi, A. Isolation and characterization of mutants affected in lipid metabolism of Chlamydomonas reinhardtii. Eur. J. Biochem. 1995, 230, 987–993. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, I.A.; Marques, S.S.I.; Cabanelas, I.T.D.; Pereira, S.A.; Druzian, J.I.; De Souza, C.O.; Vich, D.V.; De Carvalho, G.C.; Nascimento, M.A. Screening microalgae strains for biodiesel production: Lipid productivity and estimation of fuel quality based on fatty acids profiles as selective criteria. Bioenergy Res. 2013, 6, 1–13. [Google Scholar] [CrossRef]
- BenMoussa-Dahmen, I.; Chtourou, H.; Rezgui, F.; Sayadi, S.; Dhouib, A. Salinity stress increases lipid, secondary metabolites and enzyme activity in Amphora subtropica and Dunaliella sp. for biodiesel production. Bioresour. Technol. 2016, 218, 816–825. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Han, D.; Yoon, K.; Hu, Q.; Li, Y. Characterization of type 2 diacylglycerol acyltransferases in Chlamydomonas reinhardtii reveals their distinct substrate specificities and functions in triacylglycerol biosynthesis. Plant J. 2016, 86, 3–19. [Google Scholar] [CrossRef]
- Wang, T.; Ge, H.; Liu, T.; Tian, X.; Wang, Z.; Guo, M.; Chu, J.; Zhuang, Y. Salt stress induced lipid accumulation in heterotrophic culture cells of Chlorella protothecoides: Mechanisms based on the multi-level analysis of oxidative response, key enzyme activity and biochemical alteration. J. Biotechnol. 2016, 228, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Mühlroth, A.; Li, K.; Røkke, G.; Winge, P.; Olsen, Y.; Hohmann-Marriott, M.; Vadstein, O.; Bones, A. Pathways of lipid metabolism in marine algae, co-expression network, bottlenecks and candidate genes for enhanced production of EPA and DHA in species of Chromista. Mar. Drugs 2013, 11, 4662–4697. [Google Scholar] [CrossRef] [PubMed]
- Bellou, S.; Baeshen, M.N.; Elazzazy, A.M.; Aggeli, D.; Sayegh, F.; Aggelis, G. Microalgal lipids biochemistry and biotechnological perspectives. Biotechnol. Adv. 2014, 32, 1476–1493. [Google Scholar] [CrossRef]
- De Bhowmick, G.; Koduru, L.; Sen, R. Metabolic pathway engineering towards enhancing microalgal lipid biosynthesis for biofuel application-A review. Renew. Sustain. Energy Rev. 2015, 50, 1239–1253. [Google Scholar] [CrossRef]
- Campbell, W.H.; Gowri, G. Codon usage in higher plants, green algae, and cyanobacteria. Plant Physiol. 1990, 92, 1–11. [Google Scholar] [CrossRef]
- Santos-Merino, M.; Garcillán-Barcia, M.P.; De la Cruz, F. Engineering the fatty acid synthesis pathway in Synechococcus elongatus PCC 7942 improves omega-3 fatty acid production. Biotechnol. Biofuels 2018, 11, 239. [Google Scholar] [CrossRef]
- Zhang, X.; Agrawal, A.; San, K.Y. Improving fatty acid production in Escherichia coli through the overexpression of malonyl CoA-Acyl carrier protein transacylase. Biotechnol. Prog. 2012, 28, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Magnuson, K.; Oh, W.; Larson, T.J.; Cronan, J.E. Cloning and nucleotide sequence of the fabD gene encoding malonyl coenzyme A-acyl carrier protein transacylase of Escherichia coli. FEBS Lett. 1992, 299, 262–266. [Google Scholar] [CrossRef]
- Waditee-Sirisattha, R.; Kageyama, H.; Tanaka, Y.; Fukaya, M.; Takabe, T. Overexpression of halophilic serine hydroxymethyltransferase in fresh water cyanobacterium Synechococcus elongatus PCC 7942 results in increased enzyme activities of serine biosynthetic pathways and enhanced salinity tolerance. Arch. Microbiol. 2017, 199, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]





| Fatty Acid (%) (Methyl Ester) | Empty Vector | ChMAT/7942 | SynMAT/7942 |
|---|---|---|---|
| SATURATED FATTY ACIDS (SFAs) | |||
| Caprylic acid (C8:0) | 0.07 | 0.10 | 0.07 |
| Capric acid (C10:0) | 0.00 | 0.00 | 0.00 |
| Lauric acid (C12:0) | 0.00 | 0.00 | 0.00 |
| Capric acid (C10:0) | 0.00 | 0.00 | 2.09 |
| Lauric acid (C12:0) | 0.00 | 0.00 | 48.74 *** |
| Myristic acid (C14:0) | 2.07 | 2.12 | 1.12* |
| Palmitic acid (C16:0) | 42.93 | 43.52 | 0.00 |
| Stearic acid (C18:0) | 0.36 | 0.30 | 0.00 |
| Arachinodic acid (C20:0) | 0.00 | 0.00 | 0.00 |
| Behenic acid (C22:0) | 0.00 | 0.00 | 52.00 *** |
| MONO-UNSATURATED FATTY ACIDS (MUFAs) | |||
| Palmitoleic acid (C16:1) | 49.38 | 49.42 | 43.91 *** |
| Oleic acid (C18:1) | 4.99 | 4.41 | 3.80 *** |
| Erucic acid (C22:1) | 0.00 | 0.00 | 0.00 |
| Total MUFA | C58H82O5 | 53.83 | 47.71 *** |
| POLY-UNSATURATED FATTY ACIDS (PUFAs) | |||
| Linoleic acid (C18:2) | 0.20 | 0.20 | 0.29 |
| Linolenic acid (C18:3) | 0.00 | 0.00 | 0.00 |
| Total PUFA | 0.20 | 0.20 | 0.29 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Atikij, T.; Syaputri, Y.; Iwahashi, H.; Praneenararat, T.; Sirisattha, S.; Kageyama, H.; Waditee-Sirisattha, R. Enhanced Lipid Production and Molecular Dynamics under Salinity Stress in Green Microalga Chlamydomonas reinhardtii (137C). Mar. Drugs 2019, 17, 484. https://doi.org/10.3390/md17080484
Atikij T, Syaputri Y, Iwahashi H, Praneenararat T, Sirisattha S, Kageyama H, Waditee-Sirisattha R. Enhanced Lipid Production and Molecular Dynamics under Salinity Stress in Green Microalga Chlamydomonas reinhardtii (137C). Marine Drugs. 2019; 17(8):484. https://doi.org/10.3390/md17080484
Chicago/Turabian StyleAtikij, Thanapa, Yolani Syaputri, Hitoshi Iwahashi, Thanit Praneenararat, Sophon Sirisattha, Hakuto Kageyama, and Rungaroon Waditee-Sirisattha. 2019. "Enhanced Lipid Production and Molecular Dynamics under Salinity Stress in Green Microalga Chlamydomonas reinhardtii (137C)" Marine Drugs 17, no. 8: 484. https://doi.org/10.3390/md17080484
APA StyleAtikij, T., Syaputri, Y., Iwahashi, H., Praneenararat, T., Sirisattha, S., Kageyama, H., & Waditee-Sirisattha, R. (2019). Enhanced Lipid Production and Molecular Dynamics under Salinity Stress in Green Microalga Chlamydomonas reinhardtii (137C). Marine Drugs, 17(8), 484. https://doi.org/10.3390/md17080484