Probing the Anti-Cancer Potency of Sulfated Galactans on Cholangiocarcinoma Cells Using Synchrotron FTIR Microspectroscopy, Molecular Docking, and In Vitro Studies
Abstract
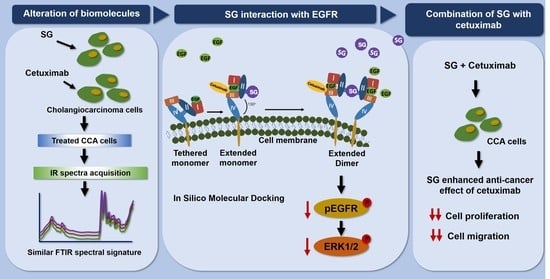
:1. Introduction
2. Results
2.1. SG and Cetuximab Inhibited CCA Cell Viability
2.2. SG, Cetuximab, and Combination of SG and Cetuximab Inhibit CCA Cell Migration
2.3. SG, Cetuximab, and Combination of SG and Cetuximab Suppressed HuCCA-1 Cell Migration by Down-Regulating Signaling Molecules in EGFR-ERK Pathway
2.4. Molecular Docking of SG with the Extracellular Domain of EGFR
2.5. SG Inhibited Ligand-Induced EGFR Dimerization
2.6. Synchrotron-FTIR Spectral Signature of HuCCA-1 Cells after Treatment with SG and Cetuximab
2.7. PCA Segregation in HuCCA-1 Cells after Treatment with SG and Cetuximab
2.8. The Biomolecules Alteration in HuCCA-1 Cells after Treatment with SG and Cetuximab
3. Discussion
4. Materials and Methods
4.1. Sulfated Galactans (SG) and Cell Lines
4.2. Effect of SG and Cetuximab on Cell Viability
4.3. Anti-Migration Effect by Wound Scratch Assay and Analysis
4.4. Western Blotting
4.5. Molecular Docking
4.6. EGFR Dimerization Assay
4.7. Synchrotron Radiation-Based Fourier-Transform Infrared Microspectroscopy (SR-FTIR-MS)
4.8. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kamsa-ard, S.; Luvira, V.; Suwanrungruang, K.; Kamsa-ard, S.; Luvira, V.; Santong, C.; Srisuk, T.; Pugkhem, A.; Bhudhisawasdi, V.; Pairojkul, C. Cholangiocarcinoma trends, incidence, and relative survival in khon kaen, thailand from 1989 through 2013:a population-based cancer registry study. J. Epidemiol. 2018, JE20180007. [Google Scholar] [CrossRef] [Green Version]
- Sripa, B.; Brindley, P.J.; Mulvenna, J.; Laha, T.; Smout, M.J.; Mairiang, E.; Bethony, J.M.; Loukas, A. The tumorigenic liver fluke Opisthorchis viverrini—Multiple pathways to cancer. Trends Parasitol. 2012, 28, 395–407. [Google Scholar] [CrossRef] [Green Version]
- Piratae, S.; Tesana, S.; Jones, M.K.; Brindley, P.J.; Loukas, A.; Lovas, E.; Eursitthichai, V.; Sripa, B.; Thanasuwan, S.; Laha, T. Molecular characterization of a tetraspanin from the human liver fluke, Opisthorchis viverrini. PLoS Negl. Trop. Dis. 2012, 6, e1939. [Google Scholar] [CrossRef] [Green Version]
- Shao, Q.; Zhu, W. Ligand binding effects on the activation of the EGFR extracellular domain. Phys. Chem. Chem. Phys. 2019, 21, 8141–8151. [Google Scholar] [CrossRef]
- Vincenzi, B.; Zoccoli, A.; Pantano, F.; Venditti, O.; Galluzzo, S. Cetuximab: From bench to bedside. Curr. Cancer Drug Targets 2010, 10, 80–95. [Google Scholar] [CrossRef]
- Li, S.; Schmitz, K.R.; Jeffrey, P.D.; Wiltzius, J.J.; Kussie, P.; Ferguson, K.M. Structural basis for inhibition of the epidermal growth factor receptor by cetuximab, cancer cell. Cancer Cell 2005, 7, 301–311. [Google Scholar] [CrossRef] [Green Version]
- Wongprasert, K.; Rudtanatip, T.; Praiboon, J. Immunostimulatory activity of sulfated galactans isolated from the red seaweed Gracilaria fisheri and development of resistance against white spot syndrome virus (WSSV) in shrimp. Fish Shellfish Immunol. 2014, 36, 52–60. [Google Scholar] [CrossRef]
- Pratoomthai, B.; Gangnonngiw, W.; Songtavisin, T. and Wongprasert, K. Effect of sulfated galactans from red seaweed Gracilaria fisheri on extracellular matrix production in human dermal fibroblast. FASEB J. 2020, 34, 1. [Google Scholar] [CrossRef]
- Rudtanatip, T.; Withyachumnarnkul, B.; Wongprasert, K. Sulfated galactans from Gracilaria fisheri bind to shrimp haemocyte membrane proteins and stimulate the expression of immune genes. Fish Shellfish Immunol. 2015, 47, 231–238. [Google Scholar] [CrossRef]
- Sae-Lao, T.; Tohtong, R.; Bates, D.O.; Wongprasert, K. Sulfated galactans from red seaweed Gracilaria fisheri target EGFR and inhibit cholangiocarcinoma cell proliferation. Am. J. Chin. Med. 2017, 45, 615–633. [Google Scholar] [CrossRef]
- Sae-Lao, T.; Luplertlop, N.; Janvilisri, T.; Tohtong, R.; Bates, D.O.; Wongprasert, K. Sulfated galactans from the red seaweed Gracilaria fisheri exerts anti-migration effect on cholangiocarcinoma cells. Phytomedicine 2017, 36, 59–67. [Google Scholar] [CrossRef]
- Bondza, S.; Foy, E.; Brooks, J.; Andersson, K.; Robinson, J.; Richalet, P.; Buijs, J. Real-time characterization of antibody binding to receptors on living immune cells. Front. Immunol. 2017, 8, 455. [Google Scholar] [CrossRef]
- Rutter, A.V.; Siddique, M.R.; Filik, J.; Sandt, C.; Dumas, P.; Cinque, G.; Sockalingum, G.D.; Yang, Y.; Sule-Suso, J. Study of gemcitabine-sensitive/resistant cancer cells by cell cloning and synchrotron FTIR microspectroscopy. Cytom. A 2014, 85, 688–697. [Google Scholar] [CrossRef]
- Wang, R.; Wang, Y. Fourier transform infrared spectroscopy in oral cancer diagnosis. Int. J. Mol. Sci 2021, 22, 1206. [Google Scholar] [CrossRef]
- Liu, Z.; Tang, Y.; Chen, F.; Liu, X.; Liu, Z.; Zhong, J.; Hu, J.; Lü, J. Synchrotron FTIR microspectroscopy reveals early adipogenic differentiation of human mesenchymal stem cells at single-cell level. Biochem. Biophys. Res. Commun. 2016, 478, 1286–1291. [Google Scholar] [CrossRef]
- Loutherback, K.; Birarda, G.; Chen, L.; Holman, H.Y. Microfluidic approaches to synchrotron radiation-based Fourier transform infrared (SR-FTIR) spectral microscopy of living biosystems. Protein. Pept. Lett. 2016, 23, 273–282. [Google Scholar] [CrossRef] [Green Version]
- Wongwattanakul, M.; Hahnvajanawong, C.; Tippayawat, P.; Chio-Srichan, S.; Leelayuwat, C.; Limpaiboon, T.; Jearanaikoon, P.; Heraud, P. Classification of gemcitabine resistant cholangiocarcinoma cell lines using synchrotron FTIR microspectroscopy. J. Biophotonics 2017, 10, 367–376. [Google Scholar] [CrossRef]
- Maphanao, P.; Thanan, R.; Loilome, W.; Chio-Srichan, S.; Wongwattanakul, M.; Sakonsinsiri, C. Synchrotron FTIR microspectroscopy revealed apoptosis-induced biomolecular changes of cholangiocarcinoma cells treated with ursolic acid. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129708. [Google Scholar] [CrossRef]
- Chatchawal, P.; Wongwattanakul, M.; Tippayawat, P.; Jearanaikoon, N.; Jumniansong, A.; Boonmars, T.; Jearanaikoon, P.; Wood, B.R. Monitoring the progression of liver fluke-induced cholangiocarcinoma in a hamster model using synchrotron FTIR microspectroscopy and focal plane array infrared imaging. Anal. Chem. 2020, 92, 15361–15369. [Google Scholar] [CrossRef]
- Yoon, J.H.; Gwak, G.Y.; Lee, H.S.; Bronk, S.F.; Werneburg, N.W.; Gores, G.J. Enhanced epidermal growth factor receptor activation in human cholangiocarcinoma cells. J. Hepatol. 2004, 41, 808–814. [Google Scholar] [CrossRef]
- Claperon, A.M.M.; Nguyen Ho-Bouldoires, T.H.; Vignjevic, D.; Wendum, D.; Chretien, Y. EGF/EGFR axis contributes to the progression of cholangiocarcinoma through the induction of an epithelial-mesenchymal transition. J. Hepatol. 2014, 61, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Dokduang, H.; Techasen, A.; Namwat, N.; Khuntikeo, N.; Pairojkul, C.; Murakami, Y.; Loilome, W.; Yongvanit, P. STATs profiling reveals predominantly-activated STAT3 in cholangiocarcinoma genesis and progression. J. Hepatobiliary Pancreat. Sci. 2014, 21, 767–776. [Google Scholar] [CrossRef] [PubMed]
- Ewald, F.; Nörz, D.; Grottke, A.; Hofmann, B.T.; Nashan, B.; Jücker, M. Dual inhibition of PI3K-AKT-mTOR- and RAF-MEK-ERK-signaling is synergistic in cholangiocarcinoma and reverses acquired resistance to MEK-inhibitors. Investig. New Drugs 2014, 32, 1144–1154. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Wu, T. Cyclooxygenase-2-derived prostaglandin E2 promotes human cholangiocarcinoma cell growth and invasion through EP1 receptor-mediated activation of the epidermal growth factor receptor and Akt. J. Biol. Chem. 2015, 290, 17806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Philip, P.A.; Benedetti, J.; Fenoglio-Preiser, C.; Zalupski, M.; Lenz, H.; O’Reilly, E.; Wong, R.; Atkins, J.; Abruzzese, J.; Blanke, C. Phase III study of gemcitabine [G] plus cetuximab [C] versus gemcitabine in patients [pts] with locally advanced or metastatic pancreatic adenocarcinoma [PC]: SWOG S0205 study. J. Clin. Oncol. 2007, 25. [Google Scholar] [CrossRef]
- Park, H.J.; Park, J.B.; Lee, S.J.; Song, M. Phellinus linteus grown on germinated brown rice increases cetuximab ssensitivity of KRAS-mutated colon cancer. Int. J. Mol. Sci. 2017, 18, 1746. [Google Scholar] [CrossRef] [Green Version]
- Schaller, M.D. Biochemical signals and biological responses elicited by the focal adhesion kinase. Biochim. Biophys. Acta 2001, 1540, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Guan, J.L. Focal adhesion kinase and its signaling pathways in cell migration and angiogenesis. Adv. Drug Deliv. Rev. 2011, 63, 610–615. [Google Scholar] [CrossRef] [Green Version]
- Eke, I.; Cordes, N. Focal adhesion signaling and therapy resistance in cancer. Semin. Cancer Biol. 2015, 31, 65–75. [Google Scholar] [CrossRef]
- Bae, G.Y.; Choi, S.J.; Lee, J.S.; Jo, J.; Lee, J.; Kim, J.; Cha, H.J. Loss of E-cadherin activates EGFR-MEK/ERK signaling, which promotes invasion via the ZEB1/MMP2 axis in non-small cell lung cancer. Oncotarget 2013, 4, 2512–2522. [Google Scholar] [CrossRef] [Green Version]
- Yan, M.D.; Yao, C.J.; Chow, J.M.; Chang, C.L.; Hwang, P.A.; Chuang, S.E.; Whang-Peng, J.; Lai, G.M. Fucoidan elevates microRNA-29b to regulate DNMT3B-MTSS1 axis and inhibit EMT in human hepatocellular carcinoma cells. Mar. Drugs 2015, 13, 6099–6116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferguson, K.M.; Berger, M.B.; Mendrola, J.M.; Cho, H.S.; Leahy, D.J.; Lemmon, M.A. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol. Cell 2003, 11, 507–517. [Google Scholar] [CrossRef]
- Harari, P.M. Epidermal growth factor receptor inhibition strategies in oncology. Endocr. Relat. Cancer 2004, 11, 689–708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabtimmai, L.; Srisook, P.; Kuaprasert, B.; Thumanu, K.; Choowongkomon, K. FTIR spectra signatures reveal different cellular effects of EGFR inhibitors on nonsmall cell lung cancer cells. J. Biophotonics 2020, 13, e201960012. [Google Scholar] [CrossRef] [PubMed]
- Greening, D.W.; Lee, S.T.; Ji, H.; Simpson, R.J.; Rigopoulos, A.; Murone, C.; Fang, C.; Gong, S.; O’Keefe, G.; Scott, A.M. Molecular profiling of cetuximab and bevacizumab treatment of colorectal tumours reveals perturbations in metabolic and hypoxic response pathways. Oncotarget 2015, 6, 38166–38180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rysman, E.; Brusselmans, K.; Scheys, K.; Timmermans, L.; Derua, R.; Munck, S.; Van Veldhoven, P.P.; Waltregny, D.; Daniels, V.W.; Machiels, J.; et al. De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer Res. 2010, 70, 8117–8126. [Google Scholar] [CrossRef] [Green Version]
- Whelan, D.R.; Bambery, K.R.; Heraud, P.; Tobin, M.J.; Diem, M.; McNaughton, D.; Wood, B.R. Monitoring the reversible B to A-like transition of DNA in eukaryotic cells using Fourier transform infrared spectroscopy. Nucleic Acids Res. 2011, 39, 5439–5448. [Google Scholar] [CrossRef] [Green Version]
- Sirisinha, S.; Tengchaisri, T.; Boonpucknavig, S.; Prempracha, N.; Ratanarapee, S.; Pausawasdi, A. Establishment and characterization of a cholangiocarcinoma cell line from a Thai patient with intrahepatic bile duct cancer. Asian Pac. J. Allergy Immunol. 1991, 9, 153–157. [Google Scholar]
- Sripa, B.; Leungwattanawanit, S.; Nitta, T.; Wongkham, C.; Bhudhisawasdi, V.; Puapairoj, A.; Sripa, C.; Miwa, M. Establishment and characterization of an opisthorchiasis-associated cholangiocarcinoma cell line (KKU-100). World J. Gastroenterol 2005, 11, 3392–3397. [Google Scholar] [CrossRef]
- Yue, P.Y.; Leung, E.P.; Mak, N.K.; Wong, R.N. A simplified method for quantifying cell migration/wound healing in 96-well plates. J. Biomol. Screen 2010, 15, 427–433. [Google Scholar] [CrossRef] [Green Version]
- Thueng-In, K.; Thanongsaksrikul, J.; Srimanote, P.; Bangphoomi, K.; Poungpair, O.; Maneewatch, S.; Choowongkomon, K.; Chaicumpa, W. Cell penetrable humanized-VH/V(H)H that inhibit RNA dependent RNA polymerase (NS5B) of HCV. PLoS ONE 2012, 7, e49254. [Google Scholar] [CrossRef] [Green Version]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol 1997, 267, 727–748. [Google Scholar] [CrossRef] [Green Version]
- Turk, H.F.; Chapkin, R.S. Analysis of epidermal growth factor receptor dimerization by BS(3) cross-linking. Methods Mol. Biol. 2015, 1233, 25–34. [Google Scholar] [CrossRef] [Green Version]
- Vivo-Truyols, G.; Schoenmakers, P.J. Automatic selection of optimal Savitzky-Golay smoothing. Anal. Chem. 2006, 78, 4598–4608. [Google Scholar] [CrossRef]








| 2nd Derivative Peak (cm−1) | Band Assignments |
|---|---|
| 2960 | -CH3 and -CH2- asymmetric stretching |
| 2923 | -CH3 and -CH2- asymmetric stretching |
| 2873 | -CH3 and -CH2- symmetric stretching |
| 2852 | -CH3 and -CH2- symmetric stretching |
| 1656 | Alpha helix of amide I |
| 1627 | Beta sheet of amide I |
| 1224, 1226, 1238, 1241, 1243 | P=O phosphodiester bond from nucleic acid |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boonsri, B.; Choowongkomon, K.; Kuaprasert, B.; Thitiphatphuvanon, T.; Supradit, K.; Sayinta, A.; Duangdara, J.; Rudtanatip, T.; Wongprasert, K. Probing the Anti-Cancer Potency of Sulfated Galactans on Cholangiocarcinoma Cells Using Synchrotron FTIR Microspectroscopy, Molecular Docking, and In Vitro Studies. Mar. Drugs 2021, 19, 258. https://doi.org/10.3390/md19050258
Boonsri B, Choowongkomon K, Kuaprasert B, Thitiphatphuvanon T, Supradit K, Sayinta A, Duangdara J, Rudtanatip T, Wongprasert K. Probing the Anti-Cancer Potency of Sulfated Galactans on Cholangiocarcinoma Cells Using Synchrotron FTIR Microspectroscopy, Molecular Docking, and In Vitro Studies. Marine Drugs. 2021; 19(5):258. https://doi.org/10.3390/md19050258
Chicago/Turabian StyleBoonsri, Boonyakorn, Kiattawee Choowongkomon, Buabarn Kuaprasert, Thanvarin Thitiphatphuvanon, Kittiya Supradit, Apinya Sayinta, Jinchutha Duangdara, Tawut Rudtanatip, and Kanokpan Wongprasert. 2021. "Probing the Anti-Cancer Potency of Sulfated Galactans on Cholangiocarcinoma Cells Using Synchrotron FTIR Microspectroscopy, Molecular Docking, and In Vitro Studies" Marine Drugs 19, no. 5: 258. https://doi.org/10.3390/md19050258
APA StyleBoonsri, B., Choowongkomon, K., Kuaprasert, B., Thitiphatphuvanon, T., Supradit, K., Sayinta, A., Duangdara, J., Rudtanatip, T., & Wongprasert, K. (2021). Probing the Anti-Cancer Potency of Sulfated Galactans on Cholangiocarcinoma Cells Using Synchrotron FTIR Microspectroscopy, Molecular Docking, and In Vitro Studies. Marine Drugs, 19(5), 258. https://doi.org/10.3390/md19050258