Isolation of Sesquiterpenoids and Steroids from the Soft Coral Sinularia brassica and Determination of Their Absolute Configuration
Abstract
:1. Introduction
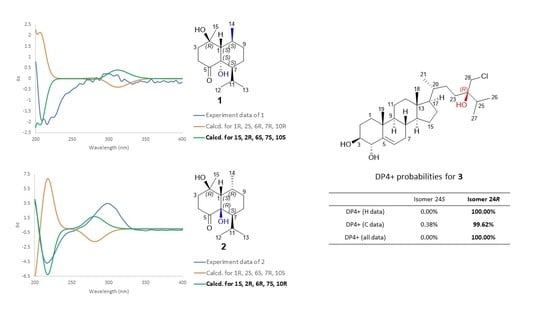
2. Results
3. Discussion
4. Materials and Methods
4.1. General Experiment Procedures
4.2. Materials and Methods
4.3. Extraction and Isolation
4.4. Computational Details
4.5. Biological Assays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Roy, P.K.; Ashimine, R.; Miyazato, H.; Taira, J.; Ueda, K. Endoperoxy and hydroperoxy cadinane-type sesquiterpenoids from an Okinawan soft coral, Sinularia sp. Arch. Pharmacal Res. 2016, 39, 778–784. [Google Scholar] [CrossRef] [PubMed]
- Su, J.H.; Huang, C.Y.; Li, P.J.; Lu, Y.; Wen, Z.H.; Kao, Y.H.; Sheu, J.H. Bioactive cadinane-type compounds from the soft coral Sinularia scabra. Arch. Pharmacal Res. 2012, 35, 779–784. [Google Scholar] [CrossRef] [PubMed]
- Lakshmi, V.; Kumar, R. Metabolites from Sinularia species. Nat. Prod. Res. 2009, 23, 801–850. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Hu, W.P.; Munro, M.H.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2009, 26, 170–244. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Liu, J.; Leng, X.; Ouyang, H. Chemical Diversity and Biological Activity of Secondary Metabolites from Soft Coral Genus Sinularia since 2013. Mar. Drugs 2021, 19, 335. [Google Scholar] [CrossRef] [PubMed]
- Eskander, R.; Al-Sofyani, A.A.; El-Sherbiny, M.M.; Ba-Akdah, M.A.; Satheesh, S. Chemical Defense of Soft Coral Sinularia polydactyla from the Red Sea Against Marine Biofilm-Forming Bacteria. J. Ocean Univ. China 2018, 17, 1451–1457. [Google Scholar] [CrossRef]
- Huang, C.Y.; Liaw, C.C.; Chen, B.W.; Chen, P.C.; Su, J.H.; Sung, P.J.; Dai, C.F.; Chiang, M.Y.; Sheu, J.H. Withanolide-based steroids from the cultured soft coral Sinularia brassica. J. Nat. Prod. 2013, 76, 1902–1908. [Google Scholar] [CrossRef]
- Tran, H.H.; Nguyen, V.P.; Nguyen Van, T.; Tran, H.T.; Nguyen Xuan, C.; Nguyen Hoai, N.; Do Cong, T.; Phan Van, K.; Chau Van, M. Cytotoxic steroid derivatives from the Vietnamese soft coral Sinularia brassica. J. Asian Nat. Prod. Res. 2017, 19, 1183–1190. [Google Scholar] [CrossRef]
- Ngoc, N.T.; Huong, P.T.; Van Thanh, N.; Cuong, N.X.; Nam, N.H.; Thung, D.C.; Van Kiem, P.; Van Minh, C. Steroid constituents from the soft coral Sinularia nanolobata. Chem. Pharm. Bull. 2016, 64, 1417–1419. [Google Scholar] [CrossRef] [Green Version]
- Quang, T.H.; Ngan, N.T.; Van Kiem, P.; Van Minh, C.; Kim, Y.H. A New Sterol from the Soft Coral Lobophytum crassum. Bull. Korean Chem. Soc. 2013, 34, 249–251. [Google Scholar]
- Rueda, A.; Zubia, E.; Ortega, M.J.; Salva, J. Structure and cytotoxicity of new polyhydroxylated sterols from the Caribbean gorgonian Plexaurella grisea. Steroids 2001, 66, 897–904. [Google Scholar] [CrossRef]
- Zhang, X.; Geoffroy, P.; Miesch, M.; Julien-David, D.; Raul, F.; Aoude-Werner, D.; Marchioni, E. Gram-scale chromatographic purification of β-sitosterol: Synthesis and characterization of β-sitosterol oxides. Steroids 2005, 70, 886–895. [Google Scholar] [CrossRef]
- Ngoc, N.T.; Huong, P.T.; Thanh, N.V.; Cuong, N.X.; Nam, N.H.; Thung, D.C.; Minh, C.V. Sesquiterpene constituents from the soft coral Sinularia nanolobata. Nat. Prod. Res. 2017, 31, 1799–1804. [Google Scholar] [CrossRef]
- Ji, N.Y.; Li, X.M.; Cui, C.M.; Wang, B.G. Terpenes and polybromoindoles from the marine red alga Laurencia decumbens (Rhodomelaceae). Helv. Chim. Acta 2007, 90, 1731–1736. [Google Scholar] [CrossRef]
- Li, R.; Shao, C.L.; Qi, X.; Li, X.B.; Li, J.; Sun, L.L.; Wang, C.Y. Polyoxygenated sterols from the South China Sea soft coral Sinularia sp. Mar. Drugs 2012, 10, 1422–1432. [Google Scholar] [CrossRef]
- Bortolotto, M.; Braekman, J.C.; Daloze, D.; Tursch, B. Chemical studies of marine invertebrates. XVIII. Four novel polyhydroxylated steroids from Sinularia dissecta (Coelenterata, Octocorallia, Alcyonacea). Bull. Sociétés Chim. Belg. 1976, 85, 27–34. [Google Scholar] [CrossRef]
- Elkhayat, E.S.; Ibrahim, S.R.; Fouad, M.A.; Mohamed, G.A. Dendronephthols A–C, new sesquiterpenoids from the Red Sea soft coral Dendronephthya sp. Tetrahedron 2014, 70, 3822–3825. [Google Scholar] [CrossRef]
- Duh, C.Y.; El-Gamal, A.A.; Song, P.Y.; Wang, S.K.; Dai, C.F. Steroids and Sesquiterpenoids from the Soft Corals Dendronephthya gigantea and Lemnalia cervicorni. J. Nat. Prod. 2004, 67, 1650–1653. [Google Scholar] [CrossRef] [PubMed]
- Xio, Y.J.; Su, J.H.; Chen, B.W.; Tseng, Y.J.; Wu, Y.C.; Sheu, J.H. Oxygenated ylangene-derived sesquiterpenoids from the soft coral Lemnalia philippinensis. Mar. Drugs 2013, 11, 3735–3741. [Google Scholar] [CrossRef] [Green Version]
- Grimblat, N.; Zanardi, M.M.; Sarotti, A.M. Beyond DP4: An improved probability for the stereochemical assignment of isomeric compounds using quantum chemical calculations of NMR shifts. J. Org. Chem. 2015, 80, 12526–12534. [Google Scholar] [CrossRef]
- Tuan, N.Q.; Oh, J.; Park, H.B.; Ferreira, D.; Choe, S.; Lee, J.; Na, M. A grayanotox-9(11)-ene derivative from Rhododendron brachycarpum and its structural assignment via a protocol combining NMR and DP4 plus application. Phytochemistry 2017, 133, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.H.; Phan, T.N.; Yoon, S.; Lee, C.J.; Jeon, H.R.; Kim, S.H.; No, J.H.; Lee, Y.S. Pyrrolidine-based 3-deoxysphingosylphosphorylcholine analogs as possible candidates against neglected tropical diseases (NTDs): Identification of hit compounds towards development of potential treatment of Leishmania donovani. J. Enzym. Inhib. Med. Chem. 2021, 36, 1922–1930. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, J.H.; Doern, G.V.; Maher, L.A.; Howell, A.W.; Redding, J.S. Antimicrobial resistance among respiratory isolates of Haemophilus influenzae, Moraxella catarrhalis, and Streptococcus pneumoniae in the United States. Antimicrob. Agents Chemother. 1990, 34, 2075–2080. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| No | δH * (J in Hz) | δC * | δH # (J in Hz) | δC # |
|---|---|---|---|---|
| 1 | 1.81 (br d, 11.0) | 51.0 | 1.55 (br d, 11.0) | 50.7 |
| 2 | - | 72.9 | - | 70.7 |
| 3 | 1.99 (m) | 39.7 | 1.80 (m) | 40.1 |
| 1.90 (ddd, 14.0, 9.6, 4.3) | 1.74 (m) | |||
| 4 | 2.75 (ddd, 15.3, 9.6, 4.7) | 35.4 | 2.52 (m) | 34.6 |
| 2.53 (ddd, 15.3, 8.9, 4.3) | 2.35 (ddd, 15.8, 6.3, 4.3) | |||
| 5 | - | 213.5 | - | 209 |
| 6 | - | 77.5 | - | 76.2 |
| 7 | 1.72 (m) | 47.9 | 1.77 (m) | 44.1 |
| 8 | 1.98 (m) | 22.8 | 1.88 (m) | 22.4 |
| 1.62 (ddt, 13.8, 4.4, 2.3) | 1.41 (ddd, 13.0, 4.3, 2.0) | |||
| 9 | 1.49 (m) | 31.2 | 1.35 (ddt, 11.9, 4.9, 2.4) | 31.2 |
| 1.30 (m) | 1.21 (m) | |||
| 10 | 2.12 (m) | 29.1 | 2.04 (m) | 28.5 |
| 11 | 1.76 (m) | 26.5 | 1.68 (m) | 26.1 |
| 12 | 0.95 (d, 6.3) | 22.5 | 0.90 (d, 6.5) | 22.7 |
| 13 | 0.72 (d, 6.5) | 23.8 | 0.66 (d, 6.8) | 23.3 |
| 14 | 1.16 (d, 6.5) | 22.7 | 1.06 (d, 6.5) | 22.6 |
| 15 | 1.43 (s) | 25.5 | 1.26 (s) | 23.1 |
| OH-2 | - | - | 4.14 (br s) | - |
| OH-6 | - | - | 4.79 (br s) | - |
| No | δH * (J in Hz) | δC * | δH # (J in Hz) | δC # |
|---|---|---|---|---|
| 1 | 2.22 (d, 4.4) | 48.6 | 1.85 (m) | 48.5 |
| 2 | - | 71.9 | - | 70.0 |
| 3 | 2.01 (m) | 40.6 | 1.75 (m) | 40.7 |
| 1.90 (m) | 1.70 (m) | |||
| 4 | 2.66 (ddd, 18.2, 8.3, 4.5) | 35.9 | 2.52 (m) | 35.2 |
| 2.60 (m) | 2.33 (ddd, 18.1, 5.7, 3.0) | |||
| 5 | - | 215.3 | - | 210.3 |
| 6 | - | 79.8 | - | 78.0 |
| 7 | 1.59 (m) | 50.8 | 1.73 (m) | 46.5 |
| 8 | 2.16 (tt, 14.2, 4.2) | 19.4 | 2.06 (m) | 18.9 |
| 1.53 (m) | 1.36 (m) | |||
| 9 | 1.82 (m) | 30.2 | 1.76 (m) | 29.9 |
| 1.36 (dq, 13.6, 3.4) | 1.20 (m) | |||
| 10 | 2.44 (m) | 28.4 | 2.39 (m) | 27.7 |
| 11 | 1.80 (m) | 26.7 | 1.74 (m) | 26.4 |
| 12 | 0.96 (d, 6.6) | 22.3 | 0.90 (d, 6.0) | 22.8 |
| 13 | 0.71 (d, 6.7) | 24.2 | 0.64 (d, 6.2) | 23.7 |
| 14 | 1.31 (d, 7.3) | 18.5 | 1.25 (d, 7.2) | 19.0 |
| 15 | 1.50 (s) | 26.1 | 1.40 (s) | 24.9 |
| OH-2 | - | - | 4.37 (s) | - |
| OH-6 | - | - | 4.64 (d, 1.9) | - |
| Isomer 24S | Isomer 24R | |
|---|---|---|
| sDP4+ (H data) | 0.25% | 99.75% |
| sDP4+ (C data) | 12.83% | 87.17% |
| sDP4+ (all data) | 0.04% | 99.96% |
| uDP4+ (H data) | 0.39% | 99.61% |
| uDP4+ (C data) | 2.53% | 97.47% |
| uDP4+ (all data) | 0.01% | 99.99% |
| DP4+ (H data) | 0.00% | 100.00% |
| DP4+ (C data) | 0.38% | 99.62% |
| DP4+ (all data) | 0.00% | 100.00% |
| No | 3 | 4 | ||
|---|---|---|---|---|
| δH (J in Hz) | δC | δH (J in Hz) | δC | |
| 1 | 1.84 (m) 1.13 (m) | 36.8 | 1.69 (m) 1.37 (m) | 32.6 |
| 2 | 1.90 (m) 1.60 (m) | 28.2 | 1.92 (m) 1.61 (m) | 31.3 |
| 3 | 3.27 (ddd, 11.3, 9.4, 4.7) | 76.7 | 3.91 (tt, 11.3, 4.8) | 68.9 |
| 4 | 4.06 (dd, 9.4, 2.7) | 75.3 | 2.07 (dd, 12.7, 11.3) 1.30 (m) | 40.0 |
| 5 | - | 142.1 | - | 65.8 |
| 6 | 5.74 (dt, 2.1, 4.9) | 117.9 | 2.90 (d, 4.4) | 59.5 |
| 7 | 2.10 (m) 1.58 (m) | 31.6 | 1.92 (m) 1.49 (dd, 15.6, 9.9) | 29.0 |
| 8 | 1.44 (m) | 31.7 | 1.36 (m) | 30.1 |
| 9 | 0.99 (m) | 50.6 | 1.25 (m) | 42.7 |
| 10 | - | 38.2 | - | 35.0 |
| 11 | 1.49 (m) | 21.0 | 1.38 (m) 1.25 (m) | 20.8 |
| 12 | 1.16 (m) 2.02 (m) | 39.8 | 1.13 (m) 1.97 (m) | 39.6 |
| 13 | - | 42.4 | - | 42.9 |
| 14 | 1.00 (m) | 56.8 | 0.96 (m) | 56.9 |
| 15 | 1.10 (m); 1.62 (m) | 24.4 | 1.00 (m); 1.58 (m) | 24.4 |
| 16 | 1.27 (m); 1.89 (m) | 28.4 | 1.28 (m); 2.00 (m) | 28.2 |
| 17 | 1.14 (m) | 55.8 | 1.20 (m) | 57.8 |
| 18 | 0.69 (s) | 12.0 | 0.59 (s) | 12.1 |
| 19 | 1.02 (s) | 20.4 | 1.06 (s) | 16.1 |
| 20 | 1.40 (m) | 36.3 | 0.98 (m) | 35.4 |
| 21 | 0.95 (overlap) | 18.9 | 0.97 (br s) | 21.2 |
| 22 | 1.42 (m) 1.07 (m) | 29.0 | 0.16(td, 8.6, 5.7) | 32.2 |
| 23 | 1.70 (m); 1.41 (m) | 31.1 | - | 25.9 |
| 24 | - | 75.4 | 0.23 (dq, 9.0, 6.9) | 50.9 |
| 25 | 1.95 (m) | 33.2 | 1.56 (m) | 32.2 |
| 26 | 0.93 (overlap) | 16.8 | 0.85 (d, 6.6) | 21.7 |
| 27 | 0.94 (overlap) | 17.2 | 0.94 (d, 6.7) | 22.3 |
| 28 | 3.56 (d, 11.2) 3.68 (d, 11.2) | 51.7 | 0.93 (d, 6.9) | 15.6 |
| 29 | - | - | 0.89 (s) | 14.4 |
| 30 | - | - | −0.14 (dd, 5.9, 4.3) 0.45 (dd, 9.1, 4.3) | 21.4 |
| Compound | Antileishmanial | Antimicrobial | |||
|---|---|---|---|---|---|
| Inhibition of Parasites (%) a | Cell Viability (%) b | C. albicans | S. aureus | E. coli | |
| IC50 (μg/mL) | |||||
| 1 | −5.3 | 96.2 | NAc | NA | NA |
| 2 | 1.4 | 96.5 | NA | NA | NA |
| 3 | 58.7 | 88.8 | NA | NA | NA |
| 4 | −11.9 | 97.1 | NA | NA | NA |
| 5 | −12.3 | 97.0 | NA | NA | NA |
| 6 | 74.3 | 106.2 | NA | >104 | NA |
| 7 | 54.7 | 96.1 | NA | NA | NA |
| 8 | 39.0 | 92.7 | NA | NA | NA |
| kanamycin | NA | 13.5 | 1.50 ± 0.24 | ||
| nystatin | 0.93 ± 0.18 | NA | NA | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pham, G.N.; Kang, D.Y.; Kim, M.J.; Han, S.J.; Lee, J.H.; Na, M. Isolation of Sesquiterpenoids and Steroids from the Soft Coral Sinularia brassica and Determination of Their Absolute Configuration. Mar. Drugs 2021, 19, 523. https://doi.org/10.3390/md19090523
Pham GN, Kang DY, Kim MJ, Han SJ, Lee JH, Na M. Isolation of Sesquiterpenoids and Steroids from the Soft Coral Sinularia brassica and Determination of Their Absolute Configuration. Marine Drugs. 2021; 19(9):523. https://doi.org/10.3390/md19090523
Chicago/Turabian StylePham, Giang Nam, Da Yeun Kang, Min Ju Kim, Se Jong Han, Jun Hyuck Lee, and MinKyun Na. 2021. "Isolation of Sesquiterpenoids and Steroids from the Soft Coral Sinularia brassica and Determination of Their Absolute Configuration" Marine Drugs 19, no. 9: 523. https://doi.org/10.3390/md19090523
APA StylePham, G. N., Kang, D. Y., Kim, M. J., Han, S. J., Lee, J. H., & Na, M. (2021). Isolation of Sesquiterpenoids and Steroids from the Soft Coral Sinularia brassica and Determination of Their Absolute Configuration. Marine Drugs, 19(9), 523. https://doi.org/10.3390/md19090523