NMR Analyses of the Enzymatic Degradation End-Products of Diabolican: The Secreted EPS of Vibrio diabolicus CNCM I-1629
Abstract
:1. Introduction
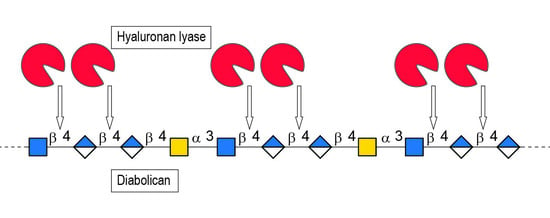
2. Results and Discussion
3. Materials and Methods
3.1. Production of the Oligosaccharides
3.2. NMR
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sutherland, I.W. Microbial polysaccharides from gram-negative bacteria. Int. Dairy J. 2001, 11, 663–674. [Google Scholar] [CrossRef]
- Freitas, F.; Alves, V.D.; Reis, M.A.M. Advances in bacterial exopolysaccharides: From production to biotechnological applications. Trends Biotechnol. 2011, 29, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Delbarre-Ladrat, C.; Sinquin, C.; Marchand, L.; Bonnetot, S.; Zykwinska, A.; Verrez-Bagnis, V.; Colliec-Jouault, S. Influence of the carbon and nitrogen sources on diabolican production by the marine Vibrio diabolicus Strain CNCM I-1629. Polymers 2022, 14, 1994. [Google Scholar] [CrossRef] [PubMed]
- Zanchetta, P.; Lagarde, N.; Guezennec, J. A new bone-healing material: A hyaluronic acid-like bacterial exopolysaccharide. Calcif. Tissue Int. 2003, 72, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Senni, K.; Gueniche, F.; Changotade, S.; Septier, D.; Sinquin, C.; Ratiskol, J.; Lutomski, D.; Godeau, G.; Guezennec, J.; Colliec-Jouault, S. Unusual glycosaminoglycans from a deep sea hydrothermal bacterium improve fibrillar collagen structuring and fibroblast activities in engineered connective tissues. Mar. Drugs 2013, 11, 1351–1369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rougeaux, H.; Kervarec, N.; Pichon, R.; Guezennec, J. Structure of the exopolysaccharide of Vibrio diabolicus isolated from a deep-sea hydrothermal vent. Carbohydr. Res. 1999, 322, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Martin-Pastor, M.; Ferreira, A.S.; Moppert, X.; Nunes, C.; Coimbra, M.A.; Reis, R.L.; Guezennec, J.; Novoa-Carballal, R. Structure, rheology, and copper-complexation of a hyaluronan-like exopolysaccharide from Vibrio. Carbohydr. Polym. 2019, 222, 114999. [Google Scholar] [CrossRef] [PubMed]
- Raguenes, G.; Christen, R.; Guezennec, J.; Pignet, P.; Barbier, G. Vibrio diabolicus sp. nov., a new polysaccharide-secreting organism isolated from a deep-sea hydrothermal vent polychaete annelid, Alvinella pompejana. Int. J. Syst. Bacteriol. 1997, 47, 989–995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rigouin, C.; Delbarre-Ladrat, C.; Ratiskol, J.; Sinquin, C.; Colliec-Jouault, S.; Dion, M. Screening of enzymatic activities for the depolymerisation of the marine bacterial exopolysaccharide HE800. Appl. Microbiol. Biotechnol. 2012, 96, 143–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helbert, W.; Poulet, L.; Drouillard, S.; Mathieu, S.; Loiodice, M.; Couturier, M.; Lombard, V.; Terrapon, N.; Turchetto, J.; Vincentelli, R.; et al. Discovery of novel carbohydrate- active enzymes through the rational exploration of the protein sequences space. Proc. Nat. Acad. Sci. USA 2019, 116, 6063–6068. [Google Scholar] [CrossRef] [PubMed]
- Esposito, F.; Vessella, G.; Sinquin, C.; Traboni, S.; Iadonisi, A.; Colliec-Jouault, S.; Zykwinska, A.; Bedini, E. Glycosaminoglycan-like sulfated polysaccharides from Vibrio diabolicus bacterium: Semi-synthesis and characterization. Carbohydr. Polym. 2022, 283, 119054. [Google Scholar] [CrossRef] [PubMed]





| Sugar Residue | 1 | 2 | 3 | 4 | 5 | 6 (6a,6b) | Ac (CH3,CO) | |
|---|---|---|---|---|---|---|---|---|
| DP3 | ||||||||
| Δ4,5-UA: D-UA-(1→ | 1H | 5.42 | 4.09 | 4.21 | 6.08 | no | no | |
| 13C | 100.89 | 69.25 | 65.55 | 108.20 | 143.20 | 168.25 | ||
| GalNAcr’α/β: →4)-α-D-GalNAcp-(1→ | 1H | 5.49/5.51 a | 4.29 | 4.01 | 4.33 | 3.96 | 3.67, 3.77 | 2.10 |
| 13C | 97.35/97.11 a | 49.62 | 67.48 | 76.15 | 70.60 | 59.95 | 21.88, 174.43 | |
| GlcNAc α: →3)-α-D-GlcNAcp | 1H | 5.20 | 4.04 | 3.95 | 3.73 | 3.91 | 3.77, 3.93 | 2.12 |
| 13C | 90.89 | 52.36 | 75.78 | 70.92 | 71.23 | 60.29 | 22.09, 174.21 | |
| GlcNAc β: →3)-β-D-GlcNAcp | 1H | 4.80 | 3.78 | 3.76 | 3.73 | 3.50 | 3.77, 3.93 | 2.11 |
| 13C | 97.47 | 55.07 | 77.92 | 70.92 | 75.65 | 60.29 | 22.09, 174.21 | |
| DP4 | ||||||||
| Δ4,5-UA: D-UA-(1→ | 1H | 5.22 | 3.95 | 4.18 | 6.07 | no | no | |
| 13C | 99.62 | 69.00 | 65.15 | 108.20 | 142.98 | 167.94 | ||
| GlcA: →4)-β-D-GlcAp-(1→ | 1H | 4.72 | 3.54 | 3.65 | 3.85 | 3.88 | no | |
| 13C | 103.07 | 73.13 | 73.92 | 79.66 | 74.82 | 173.00 | ||
| GalNAc: →4)-α-D-GalNAcp-(1→ | 1H | 5.46/5.48 a | 4.33 | 3.95 | 4.28 | 3.92 | 3.67, 3.88 | 2.10 |
| 13C | 97.28/97.04 a | 49.93/50.04 | 67.73 | 75.96 | 70.18 | 59.75 d | 21.85, c value | |
| GlcNAcrα: →3)-α-D-GlcNAcp | 1H | 5.20 | 4.05 | 3.95 | 3.73 | 3.91 | 3.77, 3.96 | 2.12 |
| 13C | 90.89 | 52.39 | 76.18 | 70.87 | 71.26 | 60.31 | 22.06, c value | |
| GlcNAcrβ: →3)-β-D-GlcNAcp | 1H | 4.79 | 3.79 | 3.76 | 3.73 | 3.52 | 3.77, 3.96 | 2.12 |
| 13C | 94.52 | 55.18 | 78.22 | 70.87 | 75.65 | 60.16 | 22.19, c value | |
| DP4 | ||||||||
| Δ4,5-UA: D-UA-(1→ | 1H | 5.41 | 4.08 | 4.21 | 6.05 | no | no | |
| 13C | 100.86 | 69.26 | 65.67 | 107.77 | 143.48 | 168.35 | ||
| GalNAc: →4)-α-D-GalNAcp-(1→ | 1H | 5.48 | 4.28 | 4.01 | 4.33 | 3.96 | 3.67, 3.88 | 2.10 |
| 13C | 97.05 | 49.57 | 67.41 | 76.09 | 70.58 | 59.82 d | 21.85, c value | |
| GlcNAc: →3)-β-D-GlcNAcp-(1→ | 1H | 4.64 | 3.84 | 3.76 | 3.73 | 3.52 | 3.77, 3.96 | 2.12 |
| 13C | 100.29/100.35 b | 53.88 | 77.98 | 70.78 | 75.55 | 60.16 | 22.43, c value | |
| GlcA α: →4)-α-D-GlcAp | 1H | 5.27 | 3.66 | 3.83 | 3.83 | 4.20 | no | |
| 13C | 91.97 | 73.92 | 70.95 | 79.84 | 71.35 | 173.00 | ||
| GlcA β: →4)-β-D-GlcAp | 1H | 4.70 | 3.37 | 3.64 | 3.83 | 3.84 | no | |
| 13C | 96.02 | 73.56 | 73.73 | 79.84 | 75.89 | 173.00 | ||
| DP7 | ||||||||
| Δ4,5-UA: D-UA-(1→ | 1H | 5.43 | 4.08 | 4.21 | 6.12 | no | no | |
| 13C | 100.88 | 69.17 | 65.48 | 108.96 | 142.54 | 167.59 | ||
| GalNAc: →4)-α-D-GalNAcp-(1→ | 1H | 5.48 | 4.28 | 4.01 | 4.32 | 3.95 | 3.67, 3.88 | 2.09 |
| 13C | 97.06 | 49.58 | 67.39 | 76.20 | 70.59 | 59.98 e | 21.85, 174.56 f | |
| GlcNAc: →3)-β-D-GlcNAcp-(1→ | 1H | 4.64 | 3.83 | 3.77 | 3.73 | 3.50 | 3.77, 3.95 | 2.10 |
| 13C | 100.42 | 53.93 | 77.98 | 70.77 | 75.55 | 60.14 | 22.41, 174.44 | |
| GlcA2: →4)-β-D-GlcAp | 1H | 4.61 | 3.42 | 3.70 | 3.85 | 3.94 | no | |
| 13C | 102.28 | 72.58 | 73.56 | 79.61 | 74.83 | 172.70 | ||
| GlcA1: →4)-β-D-GlcAp | 1H | 4.75 | 3.52 | 3.72 | 3.80 | 3.99 | no | |
| 13C | 103.24 | 72.90 | 74.08 | 80.09 | 74.29 | 173.79 | ||
| GalNAcr’: →4)-α-D-GalNAcp-(1→ | 1H | 5.46/5.48 a | 4.33 | 3.95 | 4.28 | 3.91 | 3.67, 3.88 | 2.09 |
| 13C | 97.56/97.28 a | 50.04/49.93 | 67.64/673 a | 76.16 | 70.21 | 59.86 e | 21.85, 174.53 f | |
| GlcNAcrα: →3)-α-D-GlcNAcp | 1H | 5.20 | 4.04 | 3.95 | 3.73 | 3.91 | 3.77, 3.93 | 2.10 |
| 13C | 90.89 | 52.35 | 75.69 | 70.94 | 71.36 | 60.31 | 21.96, 174.21 | |
| GlcNAcrβ: →3)-β-D-GlcNAcp | 1H | 4.79 | 3.78 | 3.76 | 3.73 | 3.50 | 3.77, 3.93 | 2.10 |
| 13C | 94.50 | 55.19 | 78.21 | 70.94 | 75.55 | 60.14 | 22.19, 174.44 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Drouillard, S.; Poulet, L.; Boisset, C.; Delbarre-Ladrat, C.; Helbert, W. NMR Analyses of the Enzymatic Degradation End-Products of Diabolican: The Secreted EPS of Vibrio diabolicus CNCM I-1629. Mar. Drugs 2022, 20, 731. https://doi.org/10.3390/md20120731
Drouillard S, Poulet L, Boisset C, Delbarre-Ladrat C, Helbert W. NMR Analyses of the Enzymatic Degradation End-Products of Diabolican: The Secreted EPS of Vibrio diabolicus CNCM I-1629. Marine Drugs. 2022; 20(12):731. https://doi.org/10.3390/md20120731
Chicago/Turabian StyleDrouillard, Sophie, Laurent Poulet, Claire Boisset, Christine Delbarre-Ladrat, and William Helbert. 2022. "NMR Analyses of the Enzymatic Degradation End-Products of Diabolican: The Secreted EPS of Vibrio diabolicus CNCM I-1629" Marine Drugs 20, no. 12: 731. https://doi.org/10.3390/md20120731
APA StyleDrouillard, S., Poulet, L., Boisset, C., Delbarre-Ladrat, C., & Helbert, W. (2022). NMR Analyses of the Enzymatic Degradation End-Products of Diabolican: The Secreted EPS of Vibrio diabolicus CNCM I-1629. Marine Drugs, 20(12), 731. https://doi.org/10.3390/md20120731