Enzyme Inhibitors from Gorgonians and Soft Corals
Abstract
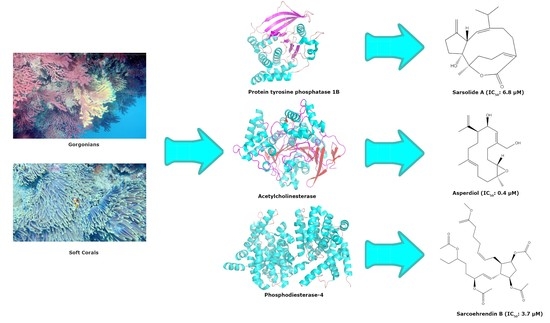
:1. Introduction
2. Enzyme Inhibitors Isolated from Gorgonians and Soft Corals
2.1. Hydrolases
2.1.1. Protein Tyrosine Phosphatase 1B (PTP1B) (EC 3.1.3.48)
2.1.2. Acetylcholinesterase (EC 3.1.1.7)
2.1.3. HIV-1 Protease (EC 3.4.23.16)
2.1.4. Elastase (EC 3.4.21.11)
2.1.5. 3CLpro Enzyme (EC 3.4.22.69)
2.1.6. Ubiquitin-Proteasome System (EC 3.4.25.1)
2.1.7. Phosphodiesterase-4 (EC 3.1.4.53)
2.1.8. α-Glucosidase (EC 3.2.1.207)
2.1.9. Histone Deacetylase 6 (HDAC6) (EC3.5.1.98)
2.1.10. Phospholipase A2 (PLA2) (EC 3.1.1.4)
2.2. Oxidoreductases
2.2.1. 5α-Reductase (EC 1.3.1.22)
2.2.2. Cytochrome P450 1A (EC 1.14.14.1)
2.2.3. Tyrosinase (EC 1.14.18.1)
2.3. Transferases
2.3.1. Tyrosine Kinase p56lck (TK) (EC 2.7.10.2)
2.3.2. IKKbeta Kinase (EC 2.7.11.10)
2.3.3. Epidermal Growth Factor Receptor Kinase (EGFR) (EC 2.7.10.1)
2.3.4. Protein Kinase C (PKC) (EC 2.7.11.13)
2.3.5. Human Tumor-Related Protein Kinases
2.3.6. Casitas B-Lineage Lymphoma Proto-Oncogene B (Cbl-b) (E3-Ubiquitin Ligase) (EC 2.3.2.27)
2.3.7. Farnesyl Protein Transferase (EC 2.5.1.58)
2.3.8. Glutathione S-Transferase (EC 2.5.1.18)
2.4. Translocases
H (+)-Pyrophosphatase (EC 7.1.3.1)
3. Walking the Whole Path to Describe an Enzyme Inhibitory Activity: What Next, after Screening and Structure Elucidation?
4. Concluding Remarks and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zakiyanov, O.; Kalousová, M.; Zima, T.; Tesař, V. Matrix Metalloproteinases and Tissue Inhibitors of Matrix Metalloproteinases in Kidney Disease. Adv. Clin. Chem. 2021, 105, 141–212. [Google Scholar] [CrossRef] [PubMed]
- Amin, F.; Khan, M.S.; Bano, B. Mammalian Cystatin and Protagonists in Brain Diseases. J. Biomol. Struct. Dyn. 2020, 38, 2171–2196. [Google Scholar] [CrossRef] [PubMed]
- Spinale, F.G.; Villarreal, F. Targeting Matrix Metalloproteinases in Heart Disease: Lessons from Endogenous Inhibitors. Biochem. Pharmacol. 2014, 90, 7–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Małgorzewicz, S.; Skrzypczak-Jankun, E.; Jankun, J. Plasminogen Activator Inhibitor-1 in Kidney Pathology (Review). Int. J. Mol. Med. 2013, 31, 503–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pini, L.; Giordani, J.; Ciarfaglia, M.; Pini, A.; Arici, M.; Tantucci, C. Alpha1-Antitrypsin Deficiency and Cardiovascular Disease: Questions and Issues of a Debated Relation. J. Cardiovasc. Med. Hagerstown Md. 2022, 23, 637–645. [Google Scholar] [CrossRef]
- Zarkadoulas, N.; Pergialiotis, V.; Dimitroulis, D.; Stefanidis, K.; Verikokos, C.; Perrea, D.N.; Kontzoglou, K. A Potential Role of Cyclin-Dependent Kinase Inhibitor 1 (P21/WAF1) in the Pathogenesis of Endometriosis: Directions for Future Research. Med. Hypotheses 2019, 133, 109414. [Google Scholar] [CrossRef]
- Grey, W.; Izatt, L.; Sahraoui, W.; Ng, Y.-M.; Ogilvie, C.; Hulse, A.; Tse, E.; Holic, R.; Yu, V. Deficiency of the Cyclin-Dependent Kinase Inhibitor, CDKN1B, Results in Overgrowth and Neurodevelopmental Delay. Hum. Mutat. 2013, 34, 864–868. [Google Scholar] [CrossRef] [Green Version]
- Sviderskaya, E.V.; Gray-Schopfer, V.C.; Hill, S.P.; Smit, N.P.; Evans-Whipp, T.J.; Bond, J.; Hill, L.; Bataille, V.; Peters, G.; Kipling, D.; et al. P16/Cyclin-Dependent Kinase Inhibitor 2A Deficiency in Human Melanocyte Senescence, Apoptosis, and Immortalization: Possible Implications for Melanoma Progression. J. Natl. Cancer Inst. 2003, 95, 723–732. [Google Scholar] [CrossRef]
- Shakeel, M.; Xu, X.; De Mandal, S.; Jin, F. Role of Serine Protease Inhibitors in Insect-Host-Pathogen Interactions. Arch. Insect Biochem. Physiol. 2019, 102, e21556. [Google Scholar] [CrossRef]
- Bao, J.; Liu, L.; An, Y.; Ran, M.; Ni, W.; Chen, J.; Wei, J.; Li, T.; Pan, G.; Zhou, Z. Nosema Bombycis Suppresses Host Hemolymph Melanization through Secreted Serpin 6 Inhibiting the Prophenoloxidase Activation Cascade. J. Invertebr. Pathol. 2019, 168, 107260. [Google Scholar] [CrossRef]
- Uemura, D.; Kawazoe, Y.; Inuzuka, T.; Itakura, Y.; Kawamata, C.; Abe, T. Drug Leads Derived from Japanese Marine Organisms. Curr. Med. Chem. 2021, 28, 196–210. [Google Scholar] [CrossRef]
- Luesch, H.; MacMillan, J.B. Targeting and Extending the Eukaryotic Druggable Genome with Natural Products. Nat. Prod. Rep. 2020, 37, 744–746. [Google Scholar] [CrossRef]
- Parvez, S.; Kang, M.; Chung, H.-S.; Bae, H. Naturally Occurring Tyrosinase Inhibitors: Mechanism and Applications in Skin Health, Cosmetics and Agriculture Industries. Phytother. Res. PTR 2007, 21, 805–816. [Google Scholar] [CrossRef]
- Mathialagan, R.; Mansor, N.; Al-Khateeb, B.; Mohamad, M.H.; Shamsuddin, M.R. Evaluation of Allicin as Soil Urease Inhibitor. Procedia Eng. 2017, 184, 449–459. [Google Scholar] [CrossRef]
- Matczuk, D.; Siczek, A. Effectiveness of the Use of Urease Inhibitors in Agriculture: A Review. Int. Agrophys. 2021, 35, 197–208. [Google Scholar] [CrossRef]
- Qiao, G.; Bi, K.; Liu, J.; Cao, S.; Liu, M.; Pešić, M.; Lin, X. Protein Kinases as Targets for Developing Anticancer Agents from Marine Organisms. Biochim. Biophys. Acta Gen. Subj. 2021, 1865, 129759. [Google Scholar] [CrossRef]
- Ning, C.; Wang, H.-M.D.; Gao, R.; Chang, Y.-C.; Hu, F.; Meng, X.; Huang, S.-Y. Marine-Derived Protein Kinase Inhibitors for Neuroinflammatory Diseases. Biomed. Eng. Online 2018, 17, 46. [Google Scholar] [CrossRef]
- Ruocco, N.; Costantini, S.; Palumbo, F.; Costantini, M. Marine Sponges and Bacteria as Challenging Sources of Enzyme Inhibitors for Pharmacological Applications. Mar. Drugs 2017, 15, 173. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Wang, N.; Zhang, T.; Zhang, B.; Sajeevan, T.P.; Joseph, V.; Armstrong, L.; He, S.; Yan, X.; Naman, C.B. A Systematic Review of Recently Reported Marine Derived Natural Product Kinase Inhibitors. Mar. Drugs 2019, 17, 493. [Google Scholar] [CrossRef] [Green Version]
- Rauf, A.; Khalil, A.A.; Olatunde, A.; Khan, M.; Anwar, S.; Alafnan, A.; Rengasamy, K.R. Diversity, Molecular Mechanisms and Structure-Activity Relationships of Marine Protease Inhibitors—A Review. Pharmacol. Res. 2021, 166, 105521. [Google Scholar] [CrossRef]
- Folmer, F.; Jaspars, M.; Schumacher, M.; Dicato, M.; Diederich, M. Marine Natural Products Targeting Phospholipases A2. Biochem. Pharmacol. 2010, 80, 1793–1800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trang, N.T.H.; Tang, D.Y.Y.; Chew, K.W.; Linh, N.T.; Hoang, L.T.; Cuong, N.T.; Yen, H.T.; Thao, N.T.; Trung, N.T.; Show, P.L.; et al. Discovery of α-Glucosidase Inhibitors from Marine Microorganisms: Optimization of Culture Conditions and Medium Composition. Mol. Biotechnol. 2021, 63, 1004–1015. [Google Scholar] [CrossRef] [PubMed]
- Lins Alves, L.K.; Cechinel Filho, V.; de Souza, R.L.R.; Furtado-Alle, L. BChE Inhibitors from Marine Organisms—A Review. Chem. Biol. Interact. 2022, 367, 110136. [Google Scholar] [CrossRef] [PubMed]
- Prasasty, V.; Radifar, M.; Istyastono, E. Natural Peptides in Drug Discovery Targeting Acetylcholinesterase. Molecules 2018, 23, 2344. [Google Scholar] [CrossRef] [Green Version]
- Dassonneville, L.; Wattez, N.; Baldeyrou, B.; Mahieu, C.; Lansiaux, A.; Banaigs, B.; Bonnard, I.; Bailly, C. Inhibition of Topoisomerase II by the Marine Alkaloid Ascididemin and Induction of Apoptosis in Leukemia Cells. Biochem. Pharmacol. 2000, 60, 527–537. [Google Scholar] [CrossRef]
- Myobatake, Y.; Takeuchi, T.; Kuramochi, K.; Kuriyama, I.; Ishido, T.; Hirano, K.; Sugawara, F.; Yoshida, H.; Mizushina, Y. Pinophilins A and B, Inhibitors of Mammalian A-, B-, and Y-Family DNA Polymerases and Human Cancer Cell Proliferation. J. Nat. Prod. 2012, 75, 135–141. [Google Scholar] [CrossRef]
- Moreno, R.I.; Zambelli, V.O.; Picolo, G.; Cury, Y.; Morandini, A.C.; Marques, A.C.; Sciani, J.M. Caspase-1 and Cathepsin B Inhibitors from Marine Invertebrates, Aiming at a Reduction in Neuroinflammation. Mar. Drugs 2022, 20, 614. [Google Scholar] [CrossRef]
- Della Sala, G.; Agriesti, F.; Mazzoccoli, C.; Tataranni, T.; Costantino, V.; Piccoli, C. Clogging the Ubiquitin-Proteasome Machinery with Marine Natural Products: Last Decade Update. Mar. Drugs 2018, 16, 467. [Google Scholar] [CrossRef] [Green Version]
- Tischler, D. A Perspective on Enzyme Inhibitors from Marine Organisms. Mar. Drugs 2020, 18, 431. [Google Scholar] [CrossRef]
- Raimundo, I.; Silva, S.G.; Costa, R.; Keller-Costa, T. Bioactive Secondary Metabolites from Octocoral-Associated Microbes-New Chances for Blue Growth. Mar. Drugs 2018, 16, 485. [Google Scholar] [CrossRef] [Green Version]
- McFadden, C.S.; Sánchez, J.A.; France, S.C. Molecular Phylogenetic Insights into the Evolution of Octocorallia: A Review. Integr. Comp. Biol. 2010, 50, 389–410. [Google Scholar] [CrossRef] [Green Version]
- Ezzat, S.M.; Bishbishy, M.H.E.; Habtemariam, S.; Salehi, B.; Sharifi-Rad, M.; Martins, N.; Sharifi-Rad, J. Looking at Marine-Derived Bioactive Molecules as Upcoming Anti-Diabetic Agents: A Special Emphasis on PTP1B Inhibitors. Molecules 2018, 23, 3334. [Google Scholar] [CrossRef] [Green Version]
- Ramírez, A.R.; Guerra, Y.; Otero, A.; García, B.; Berry, C.; Mendiola, J.; Hernández-Zanui, A.; de Los A Chávez, M. Generation of an Affinity Matrix Useful in the Purification of Natural Inhibitors of Plasmepsin II, an Antimalarial-Drug Target. Biotechnol. Appl. Biochem. 2009, 52, 149–157. [Google Scholar] [CrossRef]
- Salas-Sarduy, E.; Cabrera-Muñoz, A.; Cauerhff, A.; González-González, Y.; Trejo, S.A.; Chidichimo, A.; de Los Angeles Chávez-Planes, M.; Cazzulo, J.J. Antiparasitic Effect of a Fraction Enriched in Tight-Binding Protease Inhibitors Isolated from the Caribbean Coral Plexaura homomalla. Exp. Parasitol. 2013, 135, 611–622. [Google Scholar] [CrossRef]
- Salas-Sarduy, E.; Guerra, Y.; Covaleda Cortés, G.; Avilés, F.X.; Chávez Planes, M.A. Identification of Tight-Binding Plasmepsin II and Falcipain 2 Inhibitors in Aqueous Extracts of Marine Invertebrates by the Combination of Enzymatic and Interaction-Based Assays. Mar. Drugs 2017, 15, 123. [Google Scholar] [CrossRef] [Green Version]
- Covaleda, G.; Trejo, S.A.; Salas-Sarduy, E.; Del Rivero, M.A.; Chavez, M.A.; Aviles, F.X. Intensity Fading MALDI-TOF Mass Spectrometry and Functional Proteomics Assignments to Identify Protease Inhibitors in Marine Invertebrates. J. Proteom. 2017, 165, 75–92. [Google Scholar] [CrossRef]
- STRENDA Guidelines. Available online: https://www.beilstein-institut.de/en/projects/strenda/guidelines/ (accessed on 22 December 2022).
- Tipton, K.F.; Armstrong, R.N.; Bakker, B.M.; Bairoch, A.; Cornish-Bowden, A.; Halling, P.J.; Hofmeyr, J.-H.; Leyh, T.S.; Kettner, C.; Raushel, F.M.; et al. Standards for Reporting Enzyme Data: The STRENDA Consortium: What It Aims to Do and Why It Should Be Helpful. Perspect. Sci. 2014, 1, 131–137. [Google Scholar] [CrossRef]
- Swainston, N.; Baici, A.; Bakker, B.M.; Cornish-Bowden, A.; Fitzpatrick, P.F.; Halling, P.; Leyh, T.S.; O’Donovan, C.; Raushel, F.M.; Reschel, U.; et al. STRENDA DB: Enabling the Validation and Sharing of Enzyme Kinetics Data. FEBS J. 2018, 285, 2193–2204. [Google Scholar] [CrossRef]
- Villamar-Cruz, O.; Loza-Mejía, M.A.; Arias-Romero, L.E.; Camacho-Arroyo, I. Recent Advances in PTP1B Signaling in Metabolism and Cancer. Biosci. Rep. 2021, 41, BSR20211994. [Google Scholar] [CrossRef]
- Combs, A.P. Recent Advances in the Discovery of Competitive Protein Tyrosine Phosphatase 1B Inhibitors for the Treatment of Diabetes, Obesity, and Cancer. J. Med. Chem. 2010, 53, 2333–2344. [Google Scholar] [CrossRef]
- He, R.; Yu, Z.; Zhang, R.; Zhang, Z. Protein Tyrosine Phosphatases as Potential Therapeutic Targets. Acta Pharmacol. Sin. 2014, 35, 1227–1246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, L.F.; Kurtán, T.; Mándi, A.; Gao, L.X.; Li, J.; Zhang, W.; Guo, Y.W. Sarsolenane and Capnosane Diterpenes from the Hainan Soft Coral Sarcophyton trocheliophorum Marenzeller as PTP1B Inhibitors. Eur. J. Org. Chem. 2014, 2014, 1841–1847. [Google Scholar] [CrossRef]
- Liang, L.-F.; Gao, L.-X.; Li, J.; Taglialatela-Scafati, O.; Guo, Y.-W. Cembrane Diterpenoids from the Soft Coral Sarcophyton trocheliophorum Marenzeller as a New Class of PTP1B Inhibitors. Bioorg. Med. Chem. 2013, 21, 5076–5080. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.-F.; Kurtán, T.; Mándi, A.; Yao, L.-G.; Li, J.; Zhang, W.; Guo, Y.-W. Unprecedented Diterpenoids as a PTP1B Inhibitor from the Hainan Soft Coral Sarcophyton trocheliophorum Marenzeller. Org. Lett. 2013, 15, 274–277. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.-F.; Tang, X.-L.; Sun, Y.-T.; Luo, X.-C.; Zhang, J.; Van Ofwegen, L.; Sung, P.-J.; Li, P.-L.; Li, G.-Q. Terpenoids from the Soft Coral Sinularia sp. Collected in Yongxing Island. Mar. Drugs 2018, 16, 127. [Google Scholar] [CrossRef] [Green Version]
- Ye, F.; Zhu, Z.-D.; Gu, Y.-C.; Li, J.; Zhu, W.-L.; Guo, Y.-W. Further New Diterpenoids as PTP1B Inhibitors from the Xisha Soft Coral Sinularia polydactyla. Mar. Drugs 2018, 16, 103. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Hua, Q.; Yao, L.-G.; Liang, L.-F.; Lou, Y.-X.; Lu, Y.-H.; An, F.-L.; Guo, Y.-W. One Uncommon Bis-Sesquiterpenoid from Xisha Soft Coral Litophyton nigrum. Tetrahedron Lett. 2022, 88, 153571. [Google Scholar] [CrossRef]
- Chu, M.-J.; Tang, X.-L.; Han, X.; Li, T.; Luo, X.-C.; Jiang, M.-M.; van Ofwegen, L.; Luo, L.-Z.; Zhang, G.; Li, P.-L.; et al. Metabolites from the Paracel Islands Soft Coral Sinularia Cf. molesta. Mar. Drugs 2018, 16, 517. [Google Scholar] [CrossRef] [Green Version]
- Liang, L.-F.; Wang, X.-J.; Zhang, H.-Y.; Liu, H.-L.; Li, J.; Lan, L.-F.; Zhang, W.; Guo, Y.-W. Bioactive Polyhydroxylated Steroids from the Hainan Soft Coral Sinularia depressa Tixier-Durivault. Bioorg. Med. Chem. Lett. 2013, 23, 1334–1337. [Google Scholar] [CrossRef]
- Chen, W.-T.; Liu, H.-L.; Yao, L.-G.; Guo, Y.-W. 9,11-Secosteroids and Polyhydroxylated Steroids from Two South China Sea Soft Corals Sarcophyton trocheliophorum and Sinularia flexibilis. Steroids 2014, 92, 56–61. [Google Scholar] [CrossRef]
- Kate, A.S.; Aubry, I.; Tremblay, M.L.; Kerr, R.G. Lipidyl Pseudopteranes A-F: Isolation, Biomimetic Synthesis, and PTP1B Inhibitory Activity of a New Class of Pseudopteranoids from the Gorgonian Pseudopterogorgia acerosa. J. Nat. Prod. 2008, 71, 1977–1982. [Google Scholar] [CrossRef]
- Dos Santos, T.C.; Gomes, T.M.; Pinto, B.A.S.; Camara, A.L.; de Andrade Paes, A.M. Naturally Occurring Acetylcholinesterase Inhibitors and Their Potential Use for Alzheimer’s Disease Therapy. Front. Pharmacol. 2018, 9, 1192. [Google Scholar] [CrossRef] [Green Version]
- Haake, A.; Nguyen, K.; Friedman, L.; Chakkamparambil, B.; Grossberg, G.T. An Update on the Utility and Safety of Cholinesterase Inhibitors for the Treatment of Alzheimer’s Disease. Expert Opin. Drug Saf. 2020, 19, 147–157. [Google Scholar] [CrossRef]
- Castellanos, F.; Amaya-García, F.; Tello, E.; Ramos, F.A.; Umaña, A.; Puyana, M.; Resende, J.A.L.C.; Castellanos, L. Screening of Acetylcholinesterase Inhibitors in Marine Organisms from the Caribbean Sea. Nat. Prod. Res. 2019, 33, 3533–3540. [Google Scholar] [CrossRef]
- Ne’eman, I.; Fishelson, L.; Kashman, Y. Sarcophine—A New Toxin from the Soft Coral Sarcophyton glaucum (Alcyonaria). Toxicon 1974, 12, 593–594. [Google Scholar] [CrossRef]
- Bonnard, I.; Jhaumeer-Laulloo, S.B.; Bontemps, N.; Banaigs, B.; Aknin, M. New Lobane and Cembrane Diterpenes from Two Comorian Soft Corals. Mar. Drugs 2010, 8, 359–372. [Google Scholar] [CrossRef] [Green Version]
- Ellithey, M.S.; Lall, N.; Hussein, A.A.; Meyer, D. Cytotoxic, Cytostatic and HIV-1 PR Inhibitory Activities of the Soft Coral Litophyton arboreum. Mar. Drugs 2013, 11, 4917–4936. [Google Scholar] [CrossRef] [Green Version]
- Liao, Q.; Li, S.; Siu, S.W.I.; Yang, B.; Huang, C.; Chan, J.Y.-W.; Morlighem, J.-É.R.L.; Wong, C.T.T.; Rádis-Baptista, G.; Lee, S.M.-Y. Novel Kunitz-like Peptides Discovered in the Zoanthid Palythoa caribaeorum through Transcriptome Sequencing. J. Proteome Res. 2018, 17, 891–902. [Google Scholar] [CrossRef]
- Reina, E.; Ramos, F.A.; Castellanos, L.; Aragón, M.; Ospina, L.F. Anti-Inflammatory R-Prostaglandins from Caribbean Colombian Soft Coral Plexaura homomalla. J. Pharm. Pharmacol. 2013, 65, 1643–1652. [Google Scholar] [CrossRef]
- Avalon, N.E.; Nafie, J.; De Marco Verissimo, C.; Warrensford, L.C.; Dietrick, S.G.; Pittman, A.R.; Young, R.M.; Kearns, F.L.; Smalley, T.; Binning, J.M.; et al. Tuaimenal A, a Meroterpene from the Irish Deep-Sea Soft Coral Duva florida, Displays Inhibition of the SARS-CoV-2 3CLpro Enzyme. J. Nat. Prod. 2022, 85, 1315–1323. [Google Scholar] [CrossRef]
- Erickson, J.W. The Not-so-Great Escape. Nat. Struct. Biol. 1995, 2, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; Osswald, H.L.; Prato, G. Recent Progress in the Development of HIV-1 Protease Inhibitors for the Treatment of HIV/AIDS. J. Med. Chem. 2016, 59, 5172–5208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mocroft, A.; Lundgren, J.D. Starting Highly Active Antiretroviral Therapy: Why, When and Response to HAART. J. Antimicrob. Chemother. 2004, 54, 10–13. [Google Scholar] [CrossRef] [Green Version]
- Bardoel, B.W.; Kenny, E.F.; Sollberger, G.; Zychlinsky, A. The Balancing Act of Neutrophils. Cell Host Microbe 2014, 15, 526–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crocetti, L.; Quinn, M.T.; Schepetkin, I.A.; Giovannoni, M.P. A Patenting Perspective on Human Neutrophil Elastase (HNE) Inhibitors (2014–2018) and Their Therapeutic Applications. Expert Opin. Ther. Pat. 2019, 29, 555–578. [Google Scholar] [CrossRef]
- Luan, B.; Huynh, T.; Cheng, X.; Lan, G.; Wang, H.-R. Targeting Proteases for Treating COVID-19. J. Proteome Res. 2020, 19, 4316–4326. [Google Scholar] [CrossRef]
- Hoffman, R.L.; Kania, R.S.; Brothers, M.A.; Davies, J.F.; Ferre, R.A.; Gajiwala, K.S.; He, M.; Hogan, R.J.; Kozminski, K.; Li, L.Y.; et al. Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19. J. Med. Chem. 2020, 63, 12725–12747. [Google Scholar] [CrossRef]
- Tundo, G.R.; Sbardella, D.; Santoro, A.M.; Coletta, A.; Oddone, F.; Grasso, G.; Milardi, D.; Lacal, P.M.; Marini, S.; Purrello, R.; et al. The Proteasome as a Druggable Target with Multiple Therapeutic Potentialities: Cutting and Non-Cutting Edges. Pharmacol. Ther. 2020, 213, 107579. [Google Scholar] [CrossRef]
- Ling, X.-H.; Wang, S.-K.; Huang, Y.-H.; Huang, M.-J.; Duh, C.-Y. A High-Content Screening Assay for the Discovery of Novel Proteasome Inhibitors from Formosan Soft Corals. Mar. Drugs 2018, 16, 395. [Google Scholar] [CrossRef] [Green Version]
- Chiu, C.-Y.; Ling, X.-H.; Wang, S.-K.; Duh, C.-Y. Ubiquitin-Proteasome Modulating Dolabellanes and Secosteroids from Soft Coral Clavularia flava. Mar. Drugs 2020, 18, 39. [Google Scholar] [CrossRef] [Green Version]
- González, Y.; Doens, D.; Cruz, H.; Santamaría, R.; Gutiérrez, M.; Llanes, A.; Fernández, P.L. A Marine Diterpenoid Modulates the Proteasome Activity in Murine Macrophages Stimulated with LPS. Biomolecules 2018, 8, 109. [Google Scholar] [CrossRef] [Green Version]
- Verbitski, S.M.; Mullally, J.E.; Fitzpatrick, F.A.; Ireland, C.M. Punaglandins, Chlorinated Prostaglandins, Function as Potent Michael Receptors to Inhibit Ubiquitin Isopeptidase Activity. J. Med. Chem. 2004, 47, 2062–2070. [Google Scholar] [CrossRef]
- Cheng, Z.-B.; Deng, Y.-L.; Fan, C.-Q.; Han, Q.-H.; Lin, S.-L.; Tang, G.-H.; Luo, H.-B.; Yin, S. Prostaglandin Derivatives: Nonaromatic Phosphodiesterase-4 Inhibitors from the Soft Coral Sarcophyton ehrenbergi. J. Nat. Prod. 2014, 77, 1928–1936. [Google Scholar] [CrossRef]
- Sun, Z.-H.; Cai, Y.-H.; Fan, C.-Q.; Tang, G.-H.; Luo, H.-B.; Yin, S. Six New Tetraprenylated Alkaloids from the South China Sea Gorgonian Echinogorgia pseudossapo. Mar. Drugs 2014, 12, 672–681. [Google Scholar] [CrossRef] [Green Version]
- Mullally, J.E.; Moos, P.J.; Edes, K.; Fitzpatrick, F.A. Cyclopentenone Prostaglandins of the J Series Inhibit the Ubiquitin Isopeptidase Activity of the Proteasome Pathway. J. Biol. Chem. 2001, 276, 30366–30373. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Mansour, M.N.; Dillman, K.S.; Perez, J.R.; Danley, D.E.; Aeed, P.A.; Simons, S.P.; Lemotte, P.K.; Menniti, F.S. Structural Basis for the Catalytic Mechanism of Human Phosphodiesterase 9. Proc. Natl. Acad. Sci. USA 2008, 105, 13309–13314. [Google Scholar] [CrossRef] [Green Version]
- Burgin, A.B.; Magnusson, O.T.; Singh, J.; Witte, P.; Staker, B.L.; Bjornsson, J.M.; Thorsteinsdottir, M.; Hrafnsdottir, S.; Hagen, T.; Kiselyov, A.S.; et al. Design of Phosphodiesterase 4D (PDE4D) Allosteric Modulators for Enhancing Cognition with Improved Safety. Nat. Biotechnol. 2010, 28, 63–70. [Google Scholar] [CrossRef]
- Liu, Z.; Ma, S. Recent Advances in Synthetic α-Glucosidase Inhibitors. ChemMedChem 2017, 12, 819–829. [Google Scholar] [CrossRef]
- Asano, N. Glycosidase Inhibitors: Update and Perspectives on Practical Use. Glycobiology 2003, 13, 93R–104R. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.-J.; Wang, H.; Jiang, C.-S.; Guo, Y.-W. New Cembrane-Type Diterpenoids from the South China Sea Soft Coral Sinularia crassa and Their α-Glucosidase Inhibitory Activity. Bioorg. Chem. 2020, 104, 104281. [Google Scholar] [CrossRef]
- Andrianasolo, E.H.; Haramaty, L.; White, E.; Lutz, R.; Falkowski, P. Mode of Action of Diterpene and Characterization of Related Metabolites from the Soft Coral, Xenia elongata. Mar. Drugs 2014, 12, 1102–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, J.; Seo, Y.; Cho, K.W.; Moon, S.-S.; Cho, Y.J. Euplexides A-E: Novel Farnesylhydroquinone Glycosides from the Gorgonian Euplexaura anastomosans. J. Org. Chem. 1999, 64, 1853–1858. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.; Rho, J.R.; Cho, K.W.; Shin, J. New Farnesylhydroquinone Glycosides from the Gorgonian Euplexaura anastomosans. Nat. Prod. Lett. 2001, 15, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Seo, Y.; Rho, J.-R.; Cho, K.W. Isolation of Polyhydroxysteroids from the Gorgonian Acabaria undulata. J. Nat. Prod. 1996, 59, 679–682. [Google Scholar] [CrossRef]
- Zhang, X.; Yuan, Z.; Zhang, Y.; Yong, S.; Salas-Burgos, A.; Koomen, J.; Olashaw, N.; Parsons, J.T.; Yang, X.-J.; Dent, S.R.; et al. HDAC6 Modulates Cell Motility by Altering the Acetylation Level of Cortactin. Mol. Cell 2007, 27, 197–213. [Google Scholar] [CrossRef] [Green Version]
- Lienlaf, M.; Perez-Villarroel, P.; Knox, T.; Pabon, M.; Sahakian, E.; Powers, J.; Woan, K.V.; Lee, C.; Cheng, F.; Deng, S.; et al. Essential Role of HDAC6 in the Regulation of PD-L1 in Melanoma. Mol. Oncol. 2016, 10, 735–750. [Google Scholar] [CrossRef] [Green Version]
- Kawaguchi, Y.; Kovacs, J.J.; McLaurin, A.; Vance, J.M.; Ito, A.; Yao, T.P. The Deacetylase HDAC6 Regulates Aggresome Formation and Cell Viability in Response to Misfolded Protein Stress. Cell 2003, 115, 727–738. [Google Scholar] [CrossRef] [Green Version]
- Keremu, A.; Aimaiti, A.; Liang, Z.; Zou, X. Role of the HDAC6/STAT3 Pathway in Regulating PD-L1 Expression in Osteosarcoma Cell Lines. Cancer Chemother. Pharmacol. 2019, 83, 255–264. [Google Scholar] [CrossRef]
- Cummings, B.S. Phospholipase A2 as Targets for Anti-Cancer Drugs. Biochem. Pharmacol. 2007, 74, 949–959. [Google Scholar] [CrossRef]
- Singh, N.; Jabeen, T.; Somvanshi, R.K.; Sharma, S.; Dey, S.; Singh, T.P. Phospholipase A2 as a Target Protein for Nonsteroidal Anti-Inflammatory Drugs (NSAIDS): Crystal Structure of the Complex Formed between Phospholipase A2 and Oxyphenbutazone at 1.6 A Resolution. Biochemistry 2004, 43, 14577–14583. [Google Scholar] [CrossRef]
- Suckling, K. Phospholipase A2s: Developing Drug Targets for Atherosclerosis. Atherosclerosis 2010, 212, 357–366. [Google Scholar] [CrossRef]
- Said, M.A.; Mehta, A. The Impact of 5α-Reductase Inhibitor Use for Male Pattern Hair Loss on Men’s Health. Curr. Urol. Rep. 2018, 19, 65. [Google Scholar] [CrossRef]
- Occhiato, E.G.; Guarna, A.; Danza, G.; Serio, M. Selective Non-Steroidal Inhibitors of 5 Alpha-Reductase Type 1. J. Steroid Biochem. Mol. Biol. 2004, 88, 1–16. [Google Scholar] [CrossRef]
- Radhika, P.; Cabeza, M.; Bratoeff, E.; García, G. 5Alpha-Reductase Inhibition Activity of Steroids Isolated from Marine Soft Corals. Steroids 2004, 69, 439–444. [Google Scholar] [CrossRef]
- Liu, W.K.; Wong, N.L.Y.; Huang, H.M.; Ho, J.K.C.; Zhang, W.H.; Che, C.T. Growth Inhibitory Activity of Lemnabourside on Human Prostate Cancer Cells. Life Sci. 2002, 70, 843–853. [Google Scholar] [CrossRef]
- Hegazy, M.-E.F.; Eldeen, A.M.G.; Shahat, A.A.; Abdel-Latif, F.F.; Mohamed, T.A.; Whittlesey, B.R.; Paré, P.W. Bioactive Hydroperoxyl Cembranoids from the Red Sea Soft Coral Sarcophyton glaucum. Mar. Drugs 2012, 10, 209–222. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.-Y.S.; Wang, H.-M.D.; Wen, Y.-S.; Liu, W.; Li, P.-H.; Chiu, C.-C.; Chen, P.-C.; Huang, C.-Y.; Sheu, J.-H.; Wen, Z.-H. 4-(Phenylsulfanyl)Butan-2-One Suppresses Melanin Synthesis and Melanosome Maturation In Vitro and In Vivo. Int. J. Mol. Sci. 2015, 16, 20240–20257. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Sridhar, J.; Foroozesh, M. Cytochrome P450 Family 1 Inhibitors and Structure-Activity Relationships. Molecules 2013, 18, 14470–14495. [Google Scholar] [CrossRef] [Green Version]
- Androutsopoulos, V.P.; Tsatsakis, A.M.; Spandidos, D.A. Cytochrome P450 CYP1A1: Wider Roles in Cancer Progression and Prevention. BMC Cancer 2009, 9, 187. [Google Scholar] [CrossRef] [Green Version]
- Pillaiyar, T.; Manickam, M.; Namasivayam, V. Skin Whitening Agents: Medicinal Chemistry Perspective of Tyrosinase Inhibitors. J. Enzyme Inhib. Med. Chem. 2017, 32, 403–425. [Google Scholar] [CrossRef] [Green Version]
- Marth, J.D.; Peet, R.; Krebs, E.G.; Perlmutter, R.M. A Lymphocyte-Specific Protein-Tyrosine Kinase Gene Is Rearranged and Overexpressed in the Murine T Cell Lymphoma LSTRA. Cell 1985, 43, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Kumar Singh, P.; Kashyap, A.; Silakari, O. Exploration of the Therapeutic Aspects of Lck: A Kinase Target in Inflammatory Mediated Pathological Conditions. Biomed. Pharmacother. Biomed. Pharmacother. 2018, 108, 1565–1571. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.D.; Goclik, E.; König, G.M. Oxygenated Analogues of Gorgosterol and Ergosterol from the Soft Coral Capnella lacertiliensis. J. Nat. Prod. 2003, 66, 157–160. [Google Scholar] [CrossRef] [PubMed]
- Folmer, F.; Jaspars, M.; Solano, G.; Cristofanon, S.; Henry, E.; Tabudravu, J.; Black, K.; Green, D.H.; Küpper, F.C.; Aalbersberg, W.; et al. The Inhibition of TNF-Alpha-Induced NF-KappaB Activation by Marine Natural Products. Biochem. Pharmacol. 2009, 78, 592–606. [Google Scholar] [CrossRef] [Green Version]
- Mohyeldin, M.M.; Akl, M.R.; Siddique, A.B.; Hassan, H.M.; El Sayed, K.A. The Marine-Derived Pachycladin Diterpenoids as Novel Inhibitors of Wild-Type and Mutant EGFR. Biochem. Pharmacol. 2017, 126, 51–68. [Google Scholar] [CrossRef] [Green Version]
- He, H.; Kulanthaivel, P.; Baker, B.J.; Kalter, K.; Darges, J.; Cofield, D.; Wolff, L.; Adams, L. New Antiproliferative and Antiinflammatory 9,11-Secosterols from the Gorgonian Pseudopterogorgia sp. Tetrahedron 1995, 51, 51–58. [Google Scholar] [CrossRef]
- Zandi, E.; Rothwarf, D.M.; Delhase, M.; Hayakawa, M.; Karin, M. The IκB Kinase Complex (IKK) Contains Two Kinase Subunits, IKKα and IKKβ, Necessary for IκB Phosphorylation and NF-ΚB Activation. Cell 1997, 91, 243–252. [Google Scholar] [CrossRef] [Green Version]
- Senegas, A.; Gautheron, J.; Maurin, A.G.D.; Courtois, G. IKK-Related Genetic Diseases: Probing NF-ΚB Functions in Humans and Other Matters. Cell. Mol. Life Sci. CMLS 2015, 72, 1275–1287. [Google Scholar] [CrossRef]
- Andersson, M.; Nieuwerburgh, L.V.; Snoeijs, P. Pigment Transfer from Phytoplankton to Zooplankton with Emphasis on Astaxanthin Production in the Baltic Sea Food Web. Mar. Ecol. Prog. Ser. 2003, 254, 213–224. [Google Scholar] [CrossRef] [Green Version]
- Thompson, D.M.; Gill, G.N. The EGF Receptor: Structure, Regulation and Potential Role in Malignancy. Cancer Surv. 1985, 4, 767–788. [Google Scholar]
- Wei, Q.; Sheng, L.; Shui, Y.; Hu, Q.; Nordgren, H.; Carlsson, J. EGFR, HER2, and HER3 Expression in Laryngeal Primary Tumors and Corresponding Metastases. Ann. Surg. Oncol. 2008, 15, 1193–1201. [Google Scholar] [CrossRef]
- Wei, Q.; Chen, L.; Sheng, L.; Nordgren, H.; Wester, K.; Carlsson, J. EGFR, HER2 and HER3 Expression in Esophageal Primary Tumours and Corresponding Metastases. Int. J. Oncol. 2007, 31, 493–499. [Google Scholar] [CrossRef] [Green Version]
- Riely, G.J.; Politi, K.A.; Miller, V.A.; Pao, W. Update on Epidermal Growth Factor Receptor Mutations in Non-Small Cell Lung Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2006, 12, 7232–7241. [Google Scholar] [CrossRef] [Green Version]
- Newton, A.C. Protein Kinase C: Structural and Spatial Regulation by Phosphorylation, Cofactors, and Macromolecular Interactions. Chem. Rev. 2001, 101, 2353–2364. [Google Scholar] [CrossRef]
- Geribaldi-Doldán, N.; Gómez-Oliva, R.; Domínguez-García, S.; Nunez-Abades, P.; Castro, C. Protein Kinase C: Targets to Regenerate Brain Injuries? Front. Cell Dev. Biol. 2019, 7, 39. [Google Scholar] [CrossRef] [Green Version]
- Marrocco, V.; Bogomolovas, J.; Ehler, E.; Dos Remedios, C.G.; Yu, J.; Gao, C.; Lange, S. PKC and PKN in Heart Disease. J. Mol. Cell. Cardiol. 2019, 128, 212–226. [Google Scholar] [CrossRef] [Green Version]
- Newton, A.C. Protein Kinase C: Perfectly Balanced. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 208–230. [Google Scholar] [CrossRef] [Green Version]
- Lai, D.; Yu, S.; van Ofwegen, L.; Totzke, F.; Proksch, P.; Lin, W. 9,10-Secosteroids, Protein Kinase Inhibitors from the Chinese Gorgonian Astrogorgia sp. Bioorg. Med. Chem. 2011, 19, 6873–6880. [Google Scholar] [CrossRef]
- Zhang, J.-R.; Li, P.-L.; Tang, X.-L.; Qi, X.; Li, G.-Q. Cytotoxic Tetraprenylated Alkaloids from the South China Sea Gorgonian Euplexaura robusta. Chem. Biodivers. 2012, 9, 2218–2224. [Google Scholar] [CrossRef]
- Sorek, H.; Rudi, A.; Benayahu, Y.; Ben-Califa, N.; Neumann, D.; Kashman, Y. Nuttingins A-F and Malonganenones D-H, Tetraprenylated Alkaloids from the Tanzanian Gorgonian Euplexaura nuttingi. J. Nat. Prod. 2007, 70, 1104–1109. [Google Scholar] [CrossRef]
- Jiang, W.; Wang, D.; Wilson, B.A.P.; Voeller, D.; Bokesch, H.R.; Smith, E.A.; Lipkowitz, S.; O’Keefe, B.R.; Gustafson, K.R. Sinularamides A-G, Terpenoid-Derived Spermidine and Spermine Conjugates with Casitas B-Lineage Lymphoma Proto-Oncogene B (Cbl-b) Inhibitory Activities from a Sinularia sp. Soft Coral. J. Nat. Prod. 2021, 84, 1831–1837. [Google Scholar] [CrossRef] [PubMed]
- Coval, S.J.; Patton, R.W.; Petrin, J.M.; James, L.; Rothofsky, M.L.; Lin, S.L.; Patel, M.; Reed, J.K.; McPhail, A.T.; Bishop, W.R. A Cembranolide Diterpene Farnesyl Protein Transferase Inhibitor from the Marine Soft Coral Lobophytum cristagalli. Bioorg. Med. Chem. Lett. 1996, 6, 909–912. [Google Scholar] [CrossRef]
- Whalen, K.E.; Lane, A.L.; Kubanek, J.; Hahn, M.E. Biochemical Warfare on the Reef: The Role of Glutathione Transferases in Consumer Tolerance of Dietary Prostaglandins. PLoS ONE 2010, 5, e8537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.-C.; Gu, H. Cbl and Cbl-b in T-Cell Regulation. Trends Immunol. 2002, 23, 140–143. [Google Scholar] [CrossRef] [PubMed]
- Jafari, D.; Mousavi, M.J.; Keshavarz Shahbaz, S.; Jafarzadeh, L.; Tahmasebi, S.; Spoor, J.; Esmaeilzadeh, A. E3 Ubiquitin Ligase Casitas B Lineage Lymphoma-b and Its Potential Therapeutic Implications for Immunotherapy. Clin. Exp. Immunol. 2021, 204, 14–31. [Google Scholar] [CrossRef]
- Paolino, M.; Choidas, A.; Wallner, S.; Pranjic, B.; Uribesalgo, I.; Loeser, S.; Jamieson, A.M.; Langdon, W.Y.; Ikeda, F.; Fededa, J.P.; et al. The E3 Ligase Cbl-b and TAM Receptors Regulate Cancer Metastasis via Natural Killer Cells. Nature 2014, 507, 508–512. [Google Scholar] [CrossRef]
- Zhang, F.L.; Casey, P.J. Protein Prenylation: Molecular Mechanisms and Functional Consequences. Annu. Rev. Biochem. 1996, 65, 241–269. [Google Scholar] [CrossRef]
- Lobell, R.B.; Omer, C.A.; Abrams, M.T.; Bhimnathwala, H.G.; Brucker, M.J.; Buser, C.A.; Davide, J.P.; de Solms, S.J.; Dinsmore, C.J.; Ellis-Hutchings, M.S.; et al. Evaluation of Farnesyl:Protein Transferase and Geranylgeranyl:Protein Transferase Inhibitor Combinations in Preclinical Models. Cancer Res. 2001, 61, 8758–8768. [Google Scholar]
- Head, J.; Johnston, S.R. New Targets for Therapy in Breast Cancer: Farnesyltransferase Inhibitors. Breast Cancer Res. 2004, 6, 262. [Google Scholar] [CrossRef] [Green Version]
- Hayes, J.D.; Flanagan, J.U.; Jowsey, I.R. Glutathione Transferases. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 51–88. [Google Scholar] [CrossRef]
- O’Brien, M.L.; Tew, K.D. Glutathione and Related Enzymes in Multidrug Resistance. Eur. J. Cancer 1996, 32, 967–978. [Google Scholar] [CrossRef]
- Mahajan, S.; Atkins, W.M. The Chemistry and Biology of Inhibitors and Pro-Drugs Targeted to Glutathione S-Transferases. Cell. Mol. Life Sci. CMLS 2005, 62, 1221–1233. [Google Scholar] [CrossRef]
- Seufferheld, M.J.; Kim, K.M.; Whitfield, J.; Valerio, A.; Caetano-Anollés, G. Evolution of Vacuolar Proton Pyrophosphatase Domains and Volutin Granules: Clues into the Early Evolutionary Origin of the Acidocalcisome. Biol. Direct 2011, 6, 50. [Google Scholar] [CrossRef] [Green Version]
- Segami, S.; Asaoka, M.; Kinoshita, S.; Fukuda, M.; Nakanishi, Y.; Maeshima, M. Biochemical, Structural and Physiological Characteristics of Vacuolar H+-Pyrophosphatase. Plant Cell Physiol. 2018, 59, 1300–1308. [Google Scholar] [CrossRef]
- Hirono, M.; Ojika, M.; Mimura, H.; Nakanishi, Y.; Maeshima, M. Acylspermidine Derivatives Isolated from a Soft Coral, Sinularia sp., Inhibit Plant Vacuolar H(+)-Pyrophosphatase. J. Biochem. 2003, 133, 811–816. [Google Scholar] [CrossRef]
- Halling, P.; Fitzpatrick, P.F.; Raushel, F.M.; Rohwer, J.; Schnell, S.; Wittig, U.; Wohlgemuth, R.; Kettner, C. An Empirical Analysis of Enzyme Function Reporting for Experimental Reproducibility: Missing/Incomplete Information in Published Papers. Biophys. Chem. 2018, 242, 22–27. [Google Scholar] [CrossRef]
- Copeland, R.A. Evaluation of Enzyme Inhibitors in Drug Discovery: A Guide for Medicinal Chemists and Pharmacologists, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2013. [Google Scholar]
- Copeland, R.A. Enzymes, A Practical Introduction to Structure, Mechanism, and Data Analysis, 2nd ed.; Wiley-VCH: New York, NY, USA, 2000; ISBN 0-471-22063-9. [Google Scholar]
- Copeland, R.A. Evaluation of Enzyme Inhibitors in Drug Discovery: A Guide for Medicinal Chemists and Pharmacologists. Methods Biochem. Anal. 2005, 46, 1–265. [Google Scholar]
- Lankatillake, C.; Luo, S.; Flavel, M.; Lenon, G.B.; Gill, H.; Huynh, T.; Dias, D.A. Screening Natural Product Extracts for Potential Enzyme Inhibitors: Protocols, and the Standardisation of the Usage of Blanks in α-Amylase, α-Glucosidase and Lipase Assays. Plant Methods 2021, 17, 3. [Google Scholar] [CrossRef]
- Christopeit, T.; Øverbø, K.; Danielson, U.H.; Nilsen, I.W. Efficient Screening of Marine Extracts for Protease Inhibitors by Combining FRET Based Activity Assays and Surface Plasmon Resonance Spectroscopy Based Binding Assays. Mar. Drugs 2013, 11, 4279–4293. [Google Scholar] [CrossRef]
- Hanada, K.; Tamai, M.; Yamagishi, M.; Ohmura, S.; Sawada, J.; Tanaka, I. Isolation and Characterization of E–64, a New Thiol Protease Inhibitor. Agric. Biol. Chem. 1978, 42, 523–528. [Google Scholar] [CrossRef]
- Bennett, S.E.; Schimerlik, M.I.; Mosbaugh, D.W. Kinetics of the Uracil-DNA Glycosylase/Inhibitor Protein Association. Ung Interaction with Ugi, Nucleic Acids, and Uracil Compounds. J. Biol. Chem. 1993, 268, 26879–26885. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Jin, Y.; Wei, S.-S.; Lee, W.-H.; Zhang, Y. Purification and Characterization of an Irreversible Serine Protease Inhibitor from Skin Secretions of Bufo andrewsi. Toxicon Off. J. Int. Soc. Toxinol. 2005, 46, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Faller, B.; Bieth, J.G. Kinetics of the Interaction of Chymotrypsin with Eglin c. Biochem. J. 1991, 280 (Pt 1), 27–32. [Google Scholar] [CrossRef] [Green Version]
- Lineweaver, H.; Burk, D. The Determination of Enzyme Dissociation Constants. ACS Publ. 1934, 56, 658–666. [Google Scholar] [CrossRef]
- Dixon, M. The Determination of Enzyme Inhibitor Constants. Biochem. J. 1953, 55, 170–171. [Google Scholar] [CrossRef]
- Cortés, A.; Cascante, M.; Cárdenas, M.L.; Cornish-Bowden, A. Relationships between Inhibition Constants, Inhibitor Concentrations for 50% Inhibition and Types of Inhibition: New Ways of Analysing Data. Biochem. J. 2001, 357, 263–268. [Google Scholar] [CrossRef]
- Morrison, J.F. Kinetics of the Reversible Inhibition of Enzyme-Catalysed Reactions by Tight-Binding Inhibitors. Biochim. Biophys. Acta 1969, 185, 269–286. [Google Scholar] [CrossRef]
- Reytor Gonzalez, M.L.; Del Rivero Antigua, M.A. Reviewing the Experimental and Mathematical Factors Involved in Tight Binding Inhibitors Ki Values Determination: The Bi-Functional Protease Inhibitor SmCI as a Test Model. Biochimie 2021, 181, 86–95. [Google Scholar] [CrossRef]
- Baell, J.B.; Holloway, G.A. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J. Med. Chem. 2010, 53, 2719–2740. [Google Scholar] [CrossRef] [Green Version]
- Baell, J.B.; Nissink, J.W.M. Seven Year Itch: Pan-Assay Interference Compounds (PAINS) in 2017—Utility and Limitations. ACS Chem. Biol. 2018, 13, 36–44. [Google Scholar] [CrossRef] [Green Version]
- Baell, J.B. Feeling Nature’s PAINS: Natural Products, Natural Product Drugs, and Pan Assay Interference Compounds (PAINS). J. Nat. Prod. 2016, 79, 616–628. [Google Scholar] [CrossRef]
- Babaoglu, K.; Simeonov, A.; Irwin, J.J.; Nelson, M.E.; Feng, B.; Thomas, C.J.; Cancian, L.; Costi, M.P.; Maltby, D.A.; Jadhav, A.; et al. Comprehensive Mechanistic Analysis of Hits from High-Throughput and Docking Screens against Beta-Lactamase. J. Med. Chem. 2008, 51, 2502–2511. [Google Scholar] [CrossRef] [Green Version]
- Jadhav, A.; Ferreira, R.S.; Klumpp, C.; Mott, B.T.; Austin, C.P.; Inglese, J.; Thomas, C.J.; Maloney, D.J.; Shoichet, B.K.; Simeonov, A. Quantitative Analyses of Aggregation, Autofluorescence, and Reactivity Artifacts in a Screen for Inhibitors of a Thiol Protease. J. Med. Chem. 2010, 53, 37–51. [Google Scholar] [CrossRef] [Green Version]
- Aldrich, C.; Bertozzi, C.; Georg, G.I.; Kiessling, L.; Lindsley, C.; Liotta, D.; Merz, K.M.; Schepartz, A.; Wang, S. The Ecstasy and Agony of Assay Interference Compounds. J. Chem. Inf. Model. 2017, 57, 387–390. [Google Scholar] [CrossRef] [Green Version]
- Feng, B.Y.; Simeonov, A.; Jadhav, A.; Babaoglu, K.; Inglese, J.; Shoichet, B.K.; Austin, C.P. A High-Throughput Screen for Aggregation-Based Inhibition in a Large Compound Library. J. Med. Chem. 2007, 50, 2385–2390. [Google Scholar] [CrossRef]
- Coan, K.E.D.; Shoichet, B.K. Stoichiometry and Physical Chemistry of Promiscuous Aggregate-Based Inhibitors. J. Am. Chem. Soc. 2008, 130, 9606–9612. [Google Scholar] [CrossRef] [Green Version]











| Enzyme | EC Number | Class | Disease/Process |
|---|---|---|---|
| Protein tyrosine phosphatase 1B | 3.1.3.48 | Hydrolase | Cancer |
| Acetylcholinesterase | 3.1.1.7 | Hydrolase | Alzheimer’s disease |
| HIV-1 protease | 3.4.23.16 | Hydrolase | HIV-AIDS |
| Elastase | 3.4.21.11 | Hydrolase | Immune response/inflammation |
| 3CLpro Enzyme | 3.4.22.69 | Hydrolase | COVID-19 |
| Ubiquitin-proteasome system | 3.4.25.1 | Hydrolase | Neurodegenerative/immune-related/cancer |
| Phosphodiesterase-4 | 3.1.4.53 | Hydrolase | Inflammatory/asthma/chronic obstructive pulmonary diseases |
| α-glucosidase | 3.2.1.207 | Hydrolase | Type II diabetes |
| Histone deacetylase 6 | 3.5.1.98 | Hydrolase | Immunoregulation |
| Phospholipase A2 | 3.1.1.4 | Hydrolase | Cancer/inflammation/atherosclerosis |
| 5α-Reductase | 1.3.1.22 | Oxidoreductase | Benign hyperplasia/male pattern baldness |
| Cytochrome P450 1A | 1.14.14.1 | Oxidoreductase | Cancer |
| Tyrosinase | 1.14.18.1 | Oxidoreductase | Development of skin-whitening agents |
| Tyrosine kinase p56lck | 2.7.10.2 | Transferase | Inflammatory/autoimmune disorders |
| IKKbeta kinase | 2.7.11.10 | Transferase | Inflammatory diseases |
| Epidermal growth factor receptor kinase | 2.7.10.1 | Transferase | Cancer |
| Protein kinase C | 2.7.11.13 | Transferase | Cancer/neurological/cardiovascular disorders |
| Human tumor-related protein kinases | - | Transferase | Cancer |
| Casitas B-lineage lymphoma proto-oncogene B | 2.3.2.27 | Transferase | Cancer |
| Farnesyl protein transferase | 2.5.1.58 | Transferase | Cancer |
| Glutathione S-transferase | 2.5.1.18 | Transferase | Cancer |
| H (+)-pyrophosphatase | 7.1.3.1 | Translocase | - |
| Compound | IC50 (μM) | Source | Classification | Reference |
|---|---|---|---|---|
| Sarsolides A (1) | 6.8 | Sarcophyton trocheliophorum Marenzeller | Soft coral | [43] |
| Sarsolides B (2) | 27.1 | |||
| Sarcophytonolide N (3) | 5.95 | [44] | ||
| Sarcrassin E (4) | 6.33 | |||
| Cembrene C (5) | 26.6 | |||
| 4Z,12Z,14E-sarcophytolide (6) | 15.4 | |||
| Ketoemblide (7) | 27.2 | |||
| Methyl sarcotroates B (8) | 6.97 | [45] | ||
| Sinulin D (9) | 47,500 | Sinularia sp. | Soft coral | [46] |
| (1R,3S,4S,7E,11E)-3,4-epoxycembra-7,11,15-triene (10) | 12,500 | |||
| 15-hydroxy-α-cadinol (11) | 22,100 | |||
| Sinupol (12) | 63.9 | Sinularia polydactyla | Soft coral | [47] |
| (1R,4aR,6S,8aS)-6-((2E,4E)-6-hydroxy-6-methylhepta-2,4-dien-2-yl)-4,8a-dimethyl-1,2,4a,5,6,7,8,8a-octahyronaphthalen-1-ol (13) | 75.5 | |||
| Sinulacetate (14) | 51.8 | |||
| Linardosinene I (15) | 10.67 1 | Litophyton nigrum | Soft coral | [48] |
| Molestins C (16) | 218 | Sinularia cf. molesta | Soft coral | [49] |
| Molestins D (17) | 344 | |||
| (5′Z)-5-(2′,6′-Dimethylocta-5′,7′-dienyl)-furan-3-carboxylic acid (18) | 1.24 | |||
| (3β,4α,5α,8β)-4-methylergost-24(28)-ene-8-ol-3-monoacetate (19) | 22.7 | Sinularia depressa | Soft coral | [50] |
| (3β, 4α, 5α)-4-methylergost-24(28)-ene-3-ol (20) | 19.5 | |||
| Ergost-4,24(28)-diene-3-one (21) | 15.3 | |||
| 7α-hydroxy-crassarosterol A (22) | 33.05 | Sinularia flexibilis | Soft coral | [51] |
| Lipidyl pseudopterane A (23) | 71 1 | Pseudopterogorgia acerosa | Gorgonian | [52] |
| Lipidyl pseudopterane B (24) | ND | |||
| Lipidyl pseudopterane C (25) | ND | |||
| Lipidyl pseudopterane D (26) | ND | |||
| Lipidyl pseudopterane E (27) | ND | |||
| Lipidyl pseudopterane F (28) | ND |
| Compound | IC50 (μM) | Source | Classification | Reference |
|---|---|---|---|---|
| Acetylcholinesterase | ||||
| Asperdiol (29) | 0.358 | Eunicea knighti | Soft coral | [55] |
| Asperdiol diacetate (30) | ND | |||
| 8R-dihydroplexaurolone (31) | ND | |||
| 14-acetoxycrassine (32) | 1.40 | Pseudoplexaura porosa | Gorgonian | |
| Sarcophine (33) | ND | Sarcophyton glaucum | Soft coral | [56] |
| Sinuketal (34) | ND | Sinularia sp. | Soft coral | [46] |
| Crassumolide E (35) | ND | Lobophytum sp. | Soft coral | [57] |
| HIV-1 protease | ||||
| Alismol (36) | 7.20 | Litophyton arboreum | Soft coral | [58] |
| 7β-acetoxy-24-methylcholesta-5-24(28)-diene-3,19-diol (37) | 4.85 | |||
| erythro-N-dodecanoyl-docosasphinga-(4E,8E)-dienine (38) | 4.80 | |||
| Sarcophytol M (39) | 15.7 | |||
| Chimyl alcohol (40) | 26.6 | |||
| Elastase | ||||
| PcKuz1 | ND | Palythoa caribaeorum | Soft coral | [59] |
| PcKuz2 | ND | |||
| PcKuz3 | ND | |||
| (15R)-PGE2 (41) | ND | Plexaura homomalla | Gorgonian | [60] |
| (15R)-PGA2 (42) | ND | |||
| (15R)-OAc-PGA2 (43) | ND | |||
| 3CLpro | ||||
| Tuaimenal A (44) | 21 | Duva florida | Soft coral | [61] |
| Compound | IC50 (μM) | Source | Classification | Reference |
|---|---|---|---|---|
| Ubiquitin–Proteasome system | ||||
| Sarcophytonin A (45) | ND | Sarcophyton trocheliophorum | Soft coral | [70] |
| Laevigatol A (46) | ND | |||
| Sarcophytoxide (47) | ND | Sarcophyton ehrenbergi | Soft coral | |
| Sarcophine (33) | ND | |||
| Clavinflol C (48) | ND | Clavularia flava | Soft coral | [71] |
| Stolonidiol (49) | ND | |||
| Stolonidiol-17-acetate (50) | ND | |||
| Clavinflol B (51) | ND | |||
| 3β,11-dihydroxy-24-methyl-9,11-secocholest-5-en-9,23-dione (52) | ND | |||
| 3β,11-dihydroxy-24-methylene-9,11-secocholest-5-en-9,23-dione (53) | ND | |||
| Methyl (2R,3S,8S,9R,E)-8-hydroxy-15-methoxy-5-oxo-2,9-di(prop-1-en-2-yl)-4,14-dioxatricyclo [9.2.1.13,6] pentadeca-1(13),6,11-triene-12-carboxylate (54) | 9.77 | Pseudopterogorgia acerosa | Gorgonian | [72] |
| PNG 2 (55) | ND | Telesto riisei | Soft coral | [73] |
| PNG 3 (56) | ND | |||
| PNG 4 (57) | ND | |||
| Z-PNG 4 (58) | ND | |||
| PNG 6 (59) | ND | |||
| Phosphodiesterase-4 | ||||
| Sarcoehrendin B (60) | 3.7 | Sarcophyton ehrenbergi | Soft coral | [74] |
| Sarcoehrendin D (61) | 10.6 | |||
| Sarcoehrendin F (62) | 12.1 | |||
| Sarcoehrendin H (63) | 16.9 | |||
| Sarcoehrendin J (64) | 7.2 | |||
| 9α,15α-diacetoxy-11α-hydroxy-5Z,13E-prostadienoic acid methyl ester (65) | 4.7 | |||
| (5Z,9α,11α,13E,15S)-11,15-bis(acetoxy)-9-hydroxyprosta-5,13-dien-1-oic acid methyl ester (66) | 5.5 | |||
| 9,11,15-triacetoxy PGF2α methyl ester (67) | 1.4 | |||
| Malonganenone L (68) | 8.5 | Echinogorgia pseudossapo | Gorgonian | [75] |
| Malonganenone M (69) | ND | |||
| Malonganenone N (70) | ND | |||
| Malonganenone O (71) | ND | |||
| Malonganenone P (72) | ND | |||
| Malonganenone Q (73) | 20.3 | |||
| Compound | IC50 (μM) | Source | Classification | Reference |
|---|---|---|---|---|
| α-glucosidase | ||||
| Sinulacrassin B (74) | 10.65 | Sinularia crassa | Soft coral | [81] |
| S-(+)-cembrane A (75) | 30.31 | |||
| Histone deacetylase 6 | ||||
| Methyl (1S, 4S, 5R, 9S9-4-((Z)-4-(3,3-dimethyloxiran-2-yl-)-1-hydrioxybut-2-en-2-yl)-1-methyl-6-methylene-10-oxabicyclo[7.1.0]decane-5-carboxylate (76) | 80 | Xenia elongata | Soft coral | [82] |
| Phospholipase A2 | ||||
| Euplexide A (77) | ND | Euplexaura anastomosans | Gorgonian | [83] |
| Euplexide B (78) | ND | |||
| Euplexide F (79) | ND | [84] | ||
| Euplexide G (80) | ND | |||
| 7α,8α-epoxy-3β,5α,6α-trihydroxycholestane (81) | ND | Acabaria undulata | Gorgonian | [85] |
| 24-methyl-7α,8α-epoxy-3β,5α,6α-trihydroxycholest-22-ene (82) | ND | |||
| Compound | IC50 (μM) | Source | Classification | Reference |
|---|---|---|---|---|
| 5α-Reductase | ||||
| 24-methylenecholest-5-ene-3β,7β, 16β-triol-3-O-α-l-fucopyranoside (83) | ND | Sinularia crassa Tixier–Durivaul | Soft coral | [95] |
| Sinularia gravis Tixier–Durivault | Soft coral | |||
| Sinularia sp. | Soft coral | |||
| 24-methylenecholest-5-ene-3β,7β, 16β-diol-3-O-α-l-fucopyranoside (84) | ND | Sinularia sp. | Soft coral | |
| Cladiella sp. | Soft coral | |||
| (24S)-24-methylcholestane-3β,5α,6β,7β-tetrol (85) | ND | Lobophytum crassum | Soft coral | |
| (24S)-24-methylcholestane-3β,5α,6β,25-tetrol (86) | ND | Lobophytum sp. | Soft coral | |
| Lemnabourside (87) | 250 | Nephthea chabroli | Soft coral | [96] |
| Cytochrome P450 1A | ||||
| 12(S)-hydroperoxylsarcoph-10-ene, 8-epi-sarcophinone (88) | 2.7 | Sarcophyton glaucum | Soft coral | [97] |
| 8-epi-sarcophinone (89) | 3.7 | |||
| Ent-sarcophine (90) | 3.4 | |||
| Tyrosinase | ||||
| 4-(phenylsulfanyl)butan-2-one (91) | 34.5 * | Cladiella australis | Soft coral | [98] |
| Compound | IC50 (μM) | Source | Classification | Reference |
|---|---|---|---|---|
| Tyrosine kinase p56lck | ||||
| 12β-acetoxyergost-5-ene-3β,11β,16-triol (92) | ND | Capnella lacertiliensis | Soft coral | [104] |
| 11β-acetoxyergost-5-ene-3β,12β,16-triol (93) | ND | |||
| IKKbeta kinase | ||||
| 3,4-epoxy,13-oxo,7E,11Z,15-cembratriene (94) | ND | Sarcophyton sp. | Soft coral | [105] |
| 3,4-epoxy,13-oxo,7E,11E,15-cembratriene (95) | ND | |||
| Astaxanthin (96) | ND | Subergorgia sp. | Gorgonian | |
| EGFR kinase | ||||
| Pachycladin A (97) | ND | Cladiella pachyclados | Soft coral | [106] |
| Protein kinase C (PKC) | ||||
| 2S,4aS,5S,6S,8aS)-6-hydroxy-2-((1S,2R,3R)-3-((2R,E)-5-hydroxy-4,5,6-trimethylhept-3-en-2-yl)-2-(2-hydroxyethyl)-2-methylcyclopentyl)-5,8a-dimethyloctahydronaphthalen-1(2H)-one (98) | NA | Pseudopterogorgia sp. | Gorgonian | [107] |
| (2S,4aS,5S,6S,8aS)-6-hydroxy-2-((1S,2R,3R)-2-(2-hydroxyethyl)-2-methyl-3-((2R,5R,E)-4,5,6-trimethylhept-3-en-2-yl)cyclopentyl)-5,8a-dimethyloctahydronaphthalen-1(2H)-one (99) | NA | |||
| 3β, 11, 24-trihidroxy, 9,11-secogorgos-5-en-9-one (100) | NA | |||
| Compound | IC50 (μM) | Source | Classification | Reference |
|---|---|---|---|---|
| Human tumor-related protein kinases | ||||
| Calicoferol A (101) | 9.33 (ALK), 38.1 (Aurora-B), 21.9 (AXL), 16.9 (FAK), 3.16 (IGF-1R), 34.0 (MET wt), 2.40 (SRC), 4.95 (VEGF-R2) | Astrogorgia sp. | Gorgonian | [119] |
| Calicoferol E (102) | 4.14 (ALK), 14.7 (AXL), 9.92 (FAK), 2.42 (IGF-1R), 47.7 (MET wt), 2.24 (SRC), 4.60 (VEGF-R2) | |||
| 24-exomethylenecalicoferol E (103) | 4.36 (ALK), 20.2 (AXL), 10.7 (FAK), 2.30 (IGF-1R), 27.5 (MET wt), 1.48 (SRC), 4.85 (VEGF-R2) | |||
| 9β-hydroxy-9,10-secosteroid astrogorgol F (104) | 4.73 (ALK), 32.6 (AXL), 9.63 (FAK), 2.46 (IGF-1R), 71.5 (MET wt), 2.17 (SRC), 6.01 (VEGF-R2) | |||
| 9α-hydroxy-9,10-secosteroid astrogorgiadiol (105) | 7.55 (ALK), 25.1 (Aurora-B), 16.9 (AXL), 13.2 (FAK), 2.77 (IGF-1R), 48.9 (MEK1 wt), 78.0 (MET wt), 1.91 (SRC), 4.35 (VEGF-R2) | |||
| Malonganenone D (106) | ND | Euplexaura robusta | Gorgonian | [120] |
| Casitas B-lineage lymphoma proto-oncogene B (Cbl-b) | ||||
| Sinularamide A (107) | ND | Sinularia sp. | Soft coral | [122] |
| Sinularamide B (108) | ND | |||
| Sinularamide C (109) | 6.5 | |||
| Sinularamide D (110) | ND | |||
| Sinularamide F (111) | ND | |||
| Sinularamide G (112) | ND | |||
| Farnesyl Protein Transferase | ||||
| 1aR,4E,8E,11S,11aR,14aS,14bS)-1a,5,9,14a-tetramethyl-12-methylene-13-oxo-1a,2,3,6,7,10,11,11a,12,13,14a,14b-dodecahydrooxireno[2′,3′:13,14]cyclotetradeca[1,2-b]furan-11-yl acetate (113) | 0.15 | Lobophytum cristagalli | Soft coral | [123] |
| Glutathione S-transferase | ||||
| 15(S)-PGA2 (114) | 75.4 | Plexaura homomalla1 | Gorgonian | [124] |
| 15(R)-15-methyl PGA2 (115) | 136.7 | |||
| 15(S)-PGE2 (116) | 312.2 | |||
| 15(R)-PGE2 (117) | ND | |||
| 15(S)-PGF2a (118) | 334.6 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Córdova-Isaza, A.; Jiménez-Mármol, S.; Guerra, Y.; Salas-Sarduy, E. Enzyme Inhibitors from Gorgonians and Soft Corals. Mar. Drugs 2023, 21, 104. https://doi.org/10.3390/md21020104
Córdova-Isaza A, Jiménez-Mármol S, Guerra Y, Salas-Sarduy E. Enzyme Inhibitors from Gorgonians and Soft Corals. Marine Drugs. 2023; 21(2):104. https://doi.org/10.3390/md21020104
Chicago/Turabian StyleCórdova-Isaza, Andrea, Sofía Jiménez-Mármol, Yasel Guerra, and Emir Salas-Sarduy. 2023. "Enzyme Inhibitors from Gorgonians and Soft Corals" Marine Drugs 21, no. 2: 104. https://doi.org/10.3390/md21020104
APA StyleCórdova-Isaza, A., Jiménez-Mármol, S., Guerra, Y., & Salas-Sarduy, E. (2023). Enzyme Inhibitors from Gorgonians and Soft Corals. Marine Drugs, 21(2), 104. https://doi.org/10.3390/md21020104