Nanoparticle Enhanced Antibody and DNA Biosensors for Sensitive Detection of Salmonella
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Isolation of Salmonella Typhimurium from Human Stool Samples
2.3. Fully-Automated Microfluidic-Based Electrochemical Sensor with a New Chip Design
2.4. Sensor Chip Cleaning and SAM Deposition
2.5. Selection of HRP Concentration for Bioassays
2.6. Characterization of SAM Coated Sensor Chips Using AFM
2.7. Development of the Antibody Biosensor
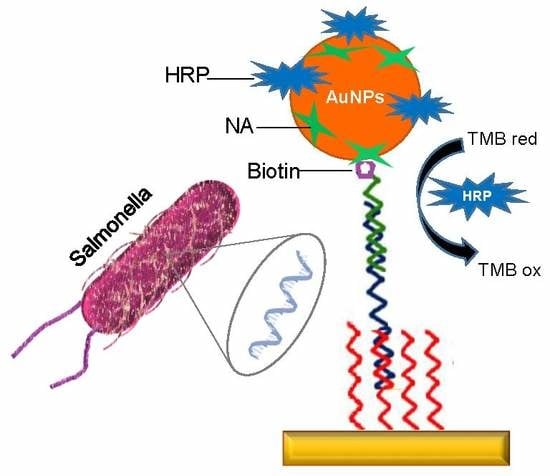
2.8. Development of the DNA Biosensor
2.8.1. Immobilization of Neutravidin and DNA Capture Probe on the Sensor Chip Surface
2.8.2. Preparation of HRP-NA-Labelled AuNPs
2.8.3. DNA Detection Assay
3. Results and Discussion
3.1. Antibody Sensor for Salmonella Detection
3.1.1. Standard Sandwich Assay for the Analysis of Commercial and Real Samples
3.1.2. Nanoparticle Enhanced Sandwich Assay for the Analysis of Commercial and Real Samples
3.1.3. Cross-Reactivity Studies for Salmonella
3.2. DNA Sensor for Salmonella Detection
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Liu, X.; Hu, Y.X.; Zheng, S.; Liu, Y.; He, Z.; Luo, F. Surface plasmon resonance immunosensor for fast, highly sensitive, and in situ detection of the magnetic nanoparticles-enriched Salmonella enteritidis. Sens. Actuators B Chem. 2016, 230, 191–198. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. National Enteric Disease Surveillance: Salmonella Annual Report; National Center for Emerging and Zoonotic Infectious Diseases, Division of Foodborne W, and Environmental Diseases: Atlanta, GA, USA, 2012.
- European Food Safety Authority (EFSA). EFSA Explains Zoonotic Diseases; EFSA: Parma, Italy, 2014. [Google Scholar]
- Ozdemir, K.; Acar, S. Plasmid profile and pulsed-field gel electrophoresis analysis of Salmonella enterica isolates from humans in Turkey. PLoS ONE 2014, 9, e95976. [Google Scholar] [CrossRef] [PubMed]
- Cinti, S.; Volpe, G.; Piermarini, S.; Delibato, E.; Palleschi, G. Electrochemical biosensors for rapid detection of foodborne Salmonella: A Critical Overview. Sensors 2017, 17, 1910. [Google Scholar] [CrossRef] [PubMed]
- Feder, I.; Nietfeld, J.C.; Galland, J.; Yeary, T.; Sargeant, J.M.; Oberst, R.; Tamplin, M.L. Comparison of cultivation and PCR-hybridization for detection of Salmonella in porcine fecal and water samples. J. Clin. Microbiol. 2001, 39, 2477–2484. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.B.; Liu, L.Q.; Song, S.S.; Tang, L.J.; Kuang, H.; Xu, C.L. A highly sensitive ELISA and immunochromatographic strip for the detection of Salmonella typhimurium in milk samples. Sensors 2015, 15, 5281–5292. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, C.; Sherwood, R.; Gheesling, L.L.; Brenner, F.W.; Fields, P.I. Molecular analysis of the rfb O antigen gene cluster of Salmonella enterica serogroup O:6,14 and development of a serogroup-specific PCR assay. Appl. Environ. Microbiol. 2003, 69, 6099–6105. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xiao, L.; Lou, Y.; Jin, M.; Liao, C.; Malakar, P.K.; Pan, Y.; Zhao, Y. Development of a multiplex real-time PCR method for simultaneous detection of Vibrio parahaemolyticus, Listeria monocytogenes and Salmonella spp. in raw shrimp. Food. Control 2015, 51, 31–36. [Google Scholar] [CrossRef]
- Maurischat, S.; Baumann, B.; Martin, A.; Malorny, B. Rapid detection and specific differentiation of Salmonella enterica subsp enterica Enteritidis, Typhimurium and its monophasic variant 4,[5], 12:i—By real-time multiplex PCR. Int. J. Food Microbiol. 2015, 193, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Altintas, Z. Biosensors and Nanotechnology: Applications in Health Care Diagnostics; Wiley: Hoboken, NJ, USA, 2017; ISBN 978-1-119-06501-2. [Google Scholar]
- Sheikhzadeh, E.; Chamsaz, M.; Turner, A.P.F.; Jager, E.W.H.; Beni, V. Label-free impedimetric biosensor for Salmonella Typhimurium detection based on poly pyrrole-co-3-carboxyl-pyrrole copolymer supported aptamer. Biosens. Bioelectron. 2016, 80, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, R.; Patel, V.; Chikkaveeraiah, B.V.; Munge, B.S.; Cheong, S.C.; Zain, R.B.; Abraham, M.T.; Dey, D.K.; Gutkind, J.S.; Rusling, J.F. Ultrasensitive detection of cancer biomarkers in the clinic by use of a nanostructured microfluidic array. Anal. Chem. 2012, 84, 6249–6255. [Google Scholar] [CrossRef] [PubMed]
- Afonso, A.S.; Uliana, C.V.; Martucci, D.H.; Faria, R.C. Simple and rapid fabrication of disposable carbon-based electrochemical cells using an electronic craft cutter for sensor and biosensor applications. Talanta 2016, 146, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Sutarlie, L.; Ow, S.Y.; Su, X.D. Nanomaterials-based biosensors for detection of microorganisms and microbial toxins. Biotechnol. J. 2017, 12, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Masdor, N.A.; Altintas, Z.; Tothill, I.E. Surface plasmon resonance immunosensor for the detection of Campylobacter jejuni. Chemosensors 2017, 5, 16. [Google Scholar] [CrossRef]
- Masdor, N.A.; Altintas, Z.; Tothill, I.E. Sensitive detection of Campylobacter jejuni using nanoparticles enhanced QCM sensor. Biosens. Bioelectron. 2016, 78, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Altintas, Z.; Akgun, M.; Kokturk, G.; Uludag, Y. A fully automated microfluidic-based electrochemical sensor for real-time bacteria detection. Biosens. Bioelectron. 2018, 100, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Melo, A.M.A.; Alexandre, D.L.; Furtado, R.F.; Borges, M.F.; Figueiredo, E.A.T.; Biswas, A.; Cheng, H.N.; Alves, C.R. Electrochemical immunosensors for Salmonella detection in food. Appl. Microbiol. Biotechnol. 2016, 100, 5301–5312. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Yan, Y.R.; Lei, P.H.; Shen, B.; Cheng, W.; Ju, H.X.; Ding, S.J. A novel electrochemical sensing strategy for rapid and ultrasensitive detection of Salmonella by rolling circle amplification and DNA-AuNPs probe. Anal. Chim. Acta 2014, 846, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Freitas, M.; Viswanathan, S.; Nouws, H.P.A.; Oliveira, M.; Delerue-Matos, C. Iron oxide/gold core/shell nanomagnetic probes and CdS biolabels for amplified electrochemical immunosensing of Salmonella typhimurium. Biosens. Bioelectron. 2014, 51, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Brandao, D.; Liebana, S.; Campoy, S.; Cortes, P.; Alegret, S.; Pividori, M.I. Electrochemical magneto-immunosensing of Salmonella based on nano and micro-sized magnetic particles. In Proceedings of the 8th Ibero-American Congress on Sensors (IBERSENSOR 2012), Carolina, Puerto Rico, 16–19 October 2012; pp. 1–7. [Google Scholar]
- Hu, C.M.; Dou, W.C.; Zhao, G.J. Enzyme immunosensor based on gold nanoparticles electroposition and Streptavidin-biotin system for detection of S. pullorum & S. gallinarum. Electrochim. Acta 2014, 117, 239–245. [Google Scholar]
- Liebana, S.; Lermo, A.; Campoy, S.; Cortes, M.P.; Alegret, S.; Pividori, M.I. Rapid detection of Salmonella in milk by electrochemical magneto-immunosensing. Biosens. Bioelectron. 2009, 25, 510–513. [Google Scholar] [CrossRef] [PubMed]
- Amini, K.; Ebralidze, I.I.; Chan, N.W.C.; Kraatz, H.-B. Characterization of TLR4/MD-2-modified Au sensor surfaces towards the detection of molecular signatures of bacteria. Anal. Methods 2016, 8, 7623–7631. [Google Scholar]
- Grimont, P.A.D.; Weill, F. Antigenic Formulae of the Salmonella Serovars, 9th ed.; WHO Collaborating Centre for Reference and Research on Salmonella; Pasteur Institute: Paris, France, 2007. [Google Scholar]
- Uludag, Y.; Esen, E.; Kokturk, G.; Ozer, H.; Muhammad, T.; Olcer, Z.; Basegmez, H.I.O.; Simsek, S.; Barut, S.; Gok, M.Y.; et al. Lab-on-a-chip based biosensor for the real-time detection of aflatoxin. Talanta 2016, 160, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Ciftci, G.Y.; Senkuytu, E.; Incir, S.E.; Yuksel, F.; Olcer, Z.; Yildirim, T.; Kilic, A.; Uludag, Y. First paraben substituted cyclotetraphosphazene compounds and DNA interaction analysis with a new automated biosensor. Biosens. Bioelectron. 2016, 80, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Altintas, Z.; Uludag, Y.; Gurbuz, Y.; Tothill, I.E. Surface plasmon resonance based immunosensor for the detection of the cancer biomarker carcinoembryonic antigen. Talanta 2011, 86, 377–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altintas, Z.; Uludag, Y.; Gurbuz, Y.; Tothill, I. Development of surface chemistry for surface plasmon resonance based sensors for the detection of proteins and DNA molecules. Anal. Chim. Acta 2012, 712, 138–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altintas, Z.; Tothill, I.E. DNA-based biosensor platforms for the detection of TP53 mutation. Sens. Actuators B Chem. 2012, 169, 188–194. [Google Scholar] [CrossRef] [Green Version]
- Pawula, M.; Altintas, Z.; Tothill, I.E. SPR detection of cardiac troponin T for acute myocardial infarction. Talanta 2016, 146, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Park, S.H.; Lee, T.H.; Nahm, B.H.; Chung, Y.H.; Seo, K.H.; Kim, H.Y. Identification of Salmonella enterica serovar Typhimurium using specific PCR primers obtained by comparative genomics in Salmonella serovars. J. Food Prot. 2006, 69, 1653–1661. [Google Scholar] [CrossRef] [PubMed]
- Altintas, Z. Surface plasmon resonance based sensor for the detection of glycopeptide antibiotics in milk using rationally designed nanoMIPs. Sci. Rep. 2018, 8, 11222. [Google Scholar] [CrossRef] [PubMed]
- Garcia, T.; Revenga-Parraa, M.; Anorga, L.; Arana, S.; Pariente, F.; Lorenzo, E. Disposable DNA biosensor based on thin-film gold electrodes for selective Salmonella detection. Sens. Actuators B Chem. 2012, 161, 1030–1037. [Google Scholar] [CrossRef]
- Zhang, D.C.; Yan, Y.R.; Li, Q.; Yu, T.X.; Cheng, W.; Wang, L.; Ju, H.X.; Ding, S.J. Label-free and high-sensitive detection of Salmonella using a surface plasmon resonance DNA-based biosensor. J. Biotechnol. 2012, 160, 123–128. [Google Scholar] [CrossRef] [PubMed]














© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Savas, S.; Ersoy, A.; Gulmez, Y.; Kilic, S.; Levent, B.; Altintas, Z. Nanoparticle Enhanced Antibody and DNA Biosensors for Sensitive Detection of Salmonella. Materials 2018, 11, 1541. https://doi.org/10.3390/ma11091541
Savas S, Ersoy A, Gulmez Y, Kilic S, Levent B, Altintas Z. Nanoparticle Enhanced Antibody and DNA Biosensors for Sensitive Detection of Salmonella. Materials. 2018; 11(9):1541. https://doi.org/10.3390/ma11091541
Chicago/Turabian StyleSavas, Sumeyra, Aylin Ersoy, Yakup Gulmez, Selcuk Kilic, Belkis Levent, and Zeynep Altintas. 2018. "Nanoparticle Enhanced Antibody and DNA Biosensors for Sensitive Detection of Salmonella" Materials 11, no. 9: 1541. https://doi.org/10.3390/ma11091541
APA StyleSavas, S., Ersoy, A., Gulmez, Y., Kilic, S., Levent, B., & Altintas, Z. (2018). Nanoparticle Enhanced Antibody and DNA Biosensors for Sensitive Detection of Salmonella. Materials, 11(9), 1541. https://doi.org/10.3390/ma11091541