Transition Metal Complexes with Flufenamic Acid for Pharmaceutical Applications—A Novel Three-Centered Coordination Polymer of Mn(II) Flufenamate
Abstract
:1. Introduction
2. Results and Discussion
2.1. Crystal Structure
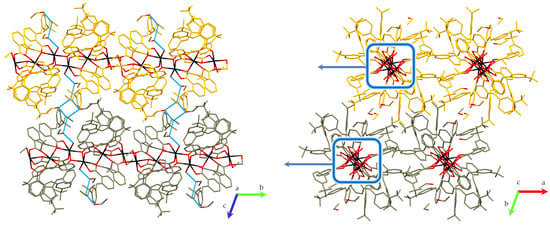
2.1.1. Description of Crystal Structure 1
2.1.2. Hirshfeld Surface Analysis of the Mn2+ Centers
2.2. Near Infrared Spectra of 1–6
2.3. TG–DTG–DTA Anaylysis
2.4. Antioxidant and Antimicrobial Activities
3. Experimental
3.1. Preparation of Complexes and Crystallization
3.2. Materials and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bagherzadeh, M.; Amini, M.; Boghaei, D.M.; Najafpour, M.M.; McKee, V. Synthesis, X–ray structure, characterization and catalytic activity of a polymeric manganese(II) complex with iminodiacetate. Appl. Organomet. Chem. 2011, 25, 559–563. [Google Scholar] [CrossRef]
- Keggin, J.F.; Miles, F.D. Structures and formulæ of the prussian blues and related compounds. Nature 1936, 137, 577–578. [Google Scholar] [CrossRef]
- Batten, S.R.; Champness, N.R. Coordination polymers and metal–organic frameworks: Materials by design. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2017, 375. [Google Scholar] [CrossRef] [Green Version]
- Ghasdian, N.; Liu, Y.; McHale, R.; He, J.; Miao, Y.; Wang, X. Synthesis of Prussian Blue Metal Coordination Polymer Nanocubes via Cyanoferrate Monomer Design. J. Inorg. Organomet. Polym. Mater. 2013, 23, 111–118. [Google Scholar] [CrossRef]
- Donaruma, L.G.; Block, B.P.; Loening, K.L.; Plate, N.; Tsuruta, T.; Buschbeck, K.C.; Powell, W.H.; Reedijk, J. Nomenclature for regular single-strand and quasi single-strand inorganic and coordination polymers. Pure Appl. Chem. 1985, 57, 149–168. [Google Scholar]
- Chen, X.M. Assembly Chemistry of Coordination Polymers. Mod. Inorg. Synth. Chem. 2011, 207–225. [Google Scholar] [CrossRef]
- Hoskins, B.F.; Robson, R. Design and Construction of a New Class of Scaffolding–like Materials Comprising Infinite Polymeric Frameworks of 3D–Linked Molecular Rods. A Reappraisal of the Zn(CN)2 and Cd(CN)2 Structures and the Synthesis and Structure of the Diamond–Related Frameworks [N(CH3)4][CuIZnII(CN)4] and CuI[4,4′,4″,4‴–tetracyanotetraphenylmethane]BF4·xC6H5NO2. J. Am. Chem. Soc. 1990, 112, 1546–1554. [Google Scholar] [CrossRef]
- Robson, R.; Abrahams, B.F.; Batten, S.R.; Gable, R.W.; Hoskins, B.F.; Liu, J. Crystal Engineering of Novel Materials Composed of Infinite Two– and Three–Dimensional Frameworks. ASC 1992, 256–273. [Google Scholar] [CrossRef]
- Wang, H.; Zhu, O.L.; Zou, R.; Xu, Q. Metal–Organic Frameworks for Energy Applications. Curr. Opin. Green Sustain. Chem. 2017, 12, 52–80. [Google Scholar] [CrossRef] [Green Version]
- Souri, B.; Rezvani, A.R.; Abbasi, S.; Hayati, P.; Janczak, J. A new Cd(II)–based coordination polymer: Conversion of morphologies from sheet–like to needle by sonochemical reaction. Inorg. Chim. Acta 2020, 509, 119692. [Google Scholar] [CrossRef]
- Ma, Z.; Moulton, B. Recent advances of discrete coordination complexes and coordination polymers in drug delivery. Coord. Chem. Rev. 2011, 255, 1623–1641. [Google Scholar] [CrossRef]
- Wang, X.K.; Liu, J.; Zhang, L.; Dong, L.Z.; Li, S.L.; Kan, Y.H.; Li, D.S.; Lan, Y.Q. Monometalic Catalytic Models Hosted in Stable Metal–Organic Frameworks for Tunable CO2 Photoreduction. ACS Catal. 2019, 9, 1726–1732. [Google Scholar] [CrossRef]
- Janiak, C. Engineering coordination polymers towards applications. Dalt. Trans. 2003, 3, 2781–2804. [Google Scholar] [CrossRef]
- Tarushi, A.; Kastanias, P.; Raptopoulou, C.P.; Psycharis, V.; Kessissoglou, D.P.; Papadopoulos, A.N.; Psomas, G. Zinc complexes of flufenamic acid: Characterization and biological evaluation. J. Inorg. Biochem. 2016, 163, 332–345. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, A.K.; Gurjar, V.; Kumar, S.; Singh, N. Molecular basis for nonspecificity of nonsteroidal anti–inflammatory drugs (NSAIDs). Drug Discov. Today 2015, 20, 863–873. [Google Scholar] [CrossRef]
- Schwier, N.; Tran, N. Non–steroidal anti–inflammatory drugs and aspirin therapy for the treatment of acute and recurrent idiopathic pericarditis. Pharmaceuticals 2016, 9, 17. [Google Scholar] [CrossRef] [Green Version]
- Sánchez–Borges, M.; Caballero–Fonseca, F.; Capriles–Hulett, A.; González–Aveledo, L. Hypersensitivity reactions to nonsteroidal anti–inflammatory drugs: An update. Pharmaceuticals 2010, 3, 10. [Google Scholar] [CrossRef]
- Ghica, M.V.; Kaya, M.G.A.; Dinu–Pîrvu, C.E.; Lupuleasa, D.; Udeanu, D.I. Development, optimization and In Vitro/In Vivo characterization of collagen–dextran spongious wound dressings loaded with flufenamic acid. Molecules 2017, 22, 1552. [Google Scholar] [CrossRef]
- Tserkezidou, C.; Hatzidimitriou, A.G.; Psomas, G. Nickel(II) complexes of flufenamic acid: Characterization, structure and interaction with DNA and albumins. Polyhedron 2016, 117, 184–192. [Google Scholar] [CrossRef]
- Guinamard, R.; Simard, C.; Del Negro, C. Flufenamic acid as an ion channel modulator. Pharmacol. Ther. 2013, 138, 272–284. [Google Scholar] [CrossRef] [Green Version]
- Mantas, A.; Mihranyan, A. Dissolution behavior of flufenamic acid in heated mixtures with nanocellulose. Molecules 2020, 25, 1277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravi, S.; Keat, A.C.; Keat, E.C. Colitis caused by non–steroidal anti–inflammatory drugs. Postgrad. Med. J. 1986, 62, 773–776. [Google Scholar] [CrossRef] [PubMed]
- Modi, C.K.; Jani, D.H. Mn(III) mixed–ligand complexes with bis–pyrazolones and ciprofloxacin drug: Synthesis, characterization and antibacterial activities. Appl. Organomet. Chem. 2011, 25, 429–436. [Google Scholar] [CrossRef]
- Lippert, B. Multiplicity of metal ion binding patterns to nucleobases. Coord. Chem. Rev. 2000, 200–202, 487–516. [Google Scholar] [CrossRef]
- Banti, C.N.; Hadjikakou, S.K. Non–Steroidal Anti–inflammatory Drugs (NSAIDs) in Metal Complexes and Their Effect at the Cellular Level. Eur. J. Inorg. Chem. 2016, 3048–3071. [Google Scholar] [CrossRef]
- Psomas, G.; Kessissoglou, D.P. Quinolones and non–steroidal anti–inflammatorydrugs interacting with copper(II), nickel(II), cobalt(II) andzinc(II): Structural features, biological evaluation and perspectives. Dalton Trans. 2013, 42, 6252–6276. [Google Scholar] [CrossRef]
- Gacki, M.; Kafarska, K.; Wolf, W.M. A supramolecular polymeric chain in the cobalt(II) complex with diclofenac: Synthesis, crystal structure, spectroscopic, thermal and antioxidant activity. J. Coord. Chem. 2019, 72, 3481–3494. [Google Scholar] [CrossRef]
- Gacki, M.; Kafarska, K.; Pietrzak, A.; Korona–Głowniak, I.; Wolf, W.M. Quasi–Isostructural Co(II) and Ni(II) Complexes with Mefenamato Ligand: Synthesis, Characterization, and Biological Activity. Molecules 2020, 25, 3099. [Google Scholar] [CrossRef]
- Kafarska, K.; Gacki, M.; Wolf, W.M. Synthesis, spectroscopic, and thermal investigations of metal complexes with mefenamic acid. J. Chem. 2017, 2017. [Google Scholar] [CrossRef] [Green Version]
- Tsiliou, S.; Kefala, L.A.; Hatzidimitriou, A.G.; Kessissoglou, D.P.; Perdih, F.; Papadopoulos, A.N.; Turel, I.; Psomas, G. Cobalt(II) complexes with non–steroidal anti–inflammatory drugs and α–diimines. J. Inorg. Biochem. 2016, 160, 125–139. [Google Scholar] [CrossRef]
- Lörinc, S.; Koman, M.; Melnik, M.; Moncoł, J.; Ondrusova, D. Bis(flufenamato–κO)bis(3–pyridylmethanol–κ2N,O)copper(II). Acta Cryst. 2004, 60, m590–m592. [Google Scholar] [CrossRef] [Green Version]
- Facchin, G.; Torre, M.H.; Kremer, E. Crystal Structure and Spectroscopic Behaviour of a Binuclear Copper(II) Complex of Flufenamic Acid and Dimethylformamide. Z. Anorg. Allg. Chem. 1998, 624, 2025–2028. [Google Scholar] [CrossRef]
- Melník, M.; Koman, M.; Glowiak, T. Structure, spectral and magnetic behaviours of tetrakis(flufenamato)(caffeine)(aqua)di copper(II)—First example of two non–equivalent ligands at the apex. Polyhedron 1998, 17, 1767–1771. [Google Scholar] [CrossRef]
- Quaresma, S.; André, V.; Fernandes, A.; Duarte, M.T. Mechanochemistry—A green synthetic methodology leading to metallodrugs, metallopharmaceuticals and bio–inspired metal–organic frameworks. Inorg. Chim. Acta 2017, 455, 309–318. [Google Scholar] [CrossRef]
- Tolia, C.; Papadopoulos, A.N.; Raptopoulou, C.P.; Psycharis, V.; Garino, C.; Salassa, L.; Psomas, G. Copper(II) interacting with the non–steroidal antiinflammatory drug flufenamic acid: Structure, antioxidant activity and binding to DNA and albumins. J. Inorg. Biochem. 2013, 123, 53–65. [Google Scholar] [CrossRef]
- Psomas, G. Copper(II) and zinc(II) coordination compounds of non–steroidal anti–inflammatory drugs: Structural features and antioxidant activity. Coord. Chem. Rev. 2020, 412, 213–259. [Google Scholar] [CrossRef]
- Smolková, R.; Zeleňák, V.; Smolko, L.; Sabolová, D.; Kuchár, J.; Gyepes, R. Novel Zn(II) complexes with non-steroidal anti-inflammatory ligand, flufenamic acid: Characterization, topoisomerase I inhibition activity, DNA and HSA binding studies. J. Inorg. Biochem. 2017, 177, 143–158. [Google Scholar] [CrossRef]
- Kumar, V.; Tewari, S.; Balendra; Karmakar, S.; Ramanan, A. Synthesis, Crystal Structures and Binding Studies of Flufenamic–Acid–Based Metal Complexes. ChemistrySelect 2018, 3, 7689–7696. [Google Scholar] [CrossRef]
- Bernstein, J.; Davis, R.; Shimoni, L.; Chang, N.-L. Patterns in hydrogen bonding: Functionality and graph set analysis in crystals. Angew. Chem. Int. Ed. 1995, 34, 1555–1573. [Google Scholar] [CrossRef]
- Etter, M.C. Encoding and decoding hydrogen–bond patterns of organic compounds. Acc. Chem. Res. 1990, 23, 120–126. [Google Scholar] [CrossRef]
- Pinto, C.B.; Dos Santos, L.H.R.; Rodrigues, B.L. Understanding metal–ligand interactions in coordination polymers using Hirshfeld surface analysis. Acta Crystallogr. Sect. C 2019, 75, 707–716. [Google Scholar] [CrossRef] [PubMed]
- Abendrot, M.; Chęcińska, L.; Kusz, J.; Lisowska, K.; Zawadzka, K.; Felczak, A.; Kalinowska–Lis, U. Zinc(II) Complexes with Amino Acids for Potential Use in Dermatology: Synthesis, Crystal Structures, and Antibacterial Activity. Molecules 2020, 25, 951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamoto, K. Infrared and Raman Spectra of Inorganic and Coordination Compounds; John Wiley & Sons, Inc.: New York, NY, USA, 2009. [Google Scholar]
- O’Donnell, F.; Smyth, T.J.P.; Ramachandran, V.N.; Smyth, W.F. A study of the antimicrobial activity of selected synthetic and naturally occurring quinolines. Int. J. Antimicrob. Agents 2009, 35, 30–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- CrysAlisPro 1.171.39.33c. Rigaku Oxford Diffraction; Agilent Technologies UK Ltd.: Yarnton, UK, 2017. [Google Scholar]
- Sheldrick, G.M. SHELXT-Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- HüBschle, C.B.; Sheldrick, G.M.; Dittrich, B. ShelXle: A Qt graphical user interface for SHELXL. J. Appl. Cryst. 2011, 44, 1281–1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Speak, A.L. PLATON SQUEEZE: A tool for the calculation of the disordered solvent contribution to the calculated structure factors. Acta Cryst. 2015, C71, 9–18. [Google Scholar]
- Macrae, C.F.; Bruno, I.J.; Chisholm, J.A.; Edgington, P.R.; Mccabe, P.; Pidcock, E.; Rodriguez–Monge, L.; Taylor, R.; Van De Streek, J.; Wood, P.A. Mercury: Visualization and analysis of crystal structures. J. Appl. Cryst. 2008, 41, 466–470. [Google Scholar] [CrossRef]
- Spek, A.L. Structure validation in chemical crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009, 65, 148–155. [Google Scholar] [CrossRef]
- Spackman, M.A.; Jayatilaka, D. Hirshfeld Surface Analysis. CrystEngComm. 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer17; University of Western Australia: Crawley, Australia, 2017. [Google Scholar]
- McKinnon, J.J.; Jayatilaka, D.; Spackman, M.A. Towards quantitative analysis of intermolecular interactions with Hirshfeld surfaces. Chem. Commun. 2007, 37, 3814–3816. [Google Scholar] [CrossRef]
- Spackman, M.A.; McKinnon, J.J. Fingerprinting intermolecular interactions in molecular crystals. CrystEngComm 2002, 4, 378–392. [Google Scholar] [CrossRef]






| No. | Compounds | Empirical Formula | Color | Formula Weight [g/mol] | Elemental Analysis, Found (calc.) [%] | |||
|---|---|---|---|---|---|---|---|---|
| C | H | N | M * | |||||
| 1 | [Mn3(fluf)6(H2O)(EtOH)]·3EtOH | C92H80F18Mn3N6O17 | Pale pink | 2048.44 | 54.21 (53.94) | 3.87 (3.94) | 4.67 (4.10) | 8.36 (8.05) |
| 2 | [Co(fluf)2(EtOH)(H2O)]·H2O | C30H28F6CoN2O7 | Pink | 701.48 | 51.68 (51.37) | 3.51 (4.02) | 3.77 (3.99) | 8.78 (8.40) |
| 3 | [Ni(fluf)2(EtOH)(H2O)]·H2O | C30H28F6NiN2O7 | Green | 701.24 | 51.88 (51.38) | 3.61 (4.02) | 3.67 (3.99) | 8.18 (8.37) |
| 4 | [Cu(fluf)2H2O] | C28H20F6CuN2O5 | Green | 642.02 | 52.21 (52.38) | 3.87 (3.14) | 4.27 (4.36) | 9.71 (9.90) |
| 5 | [Zn(fluf)2(H2O)] | C28H20F6ZnN2O5 | White | 643.86 | 52.21 (52.23) | 3.02 (3.13) | 4.17 (4.35) | 10.18 (10.15) |
| 1 | |
|---|---|
| T (K) | 100.0 |
| Crystal system | triclinic |
| Space group | P |
| a (Å) | 15.2886 (3) |
| b (Å) | 17.4001 (4) |
| c (Å) | 18.0882 (4) |
| α (°) | 98.841 (2) |
| β (°) | 93.192 (2) |
| γ (°) | 108.922 (2) |
| V (Å3) | 4469.01 (18) |
| Z | 2 |
| ρcalc (g/cm3) | 1.495 |
| F(000) | 2052 |
| Radiation | MoKα (λ = 0.71073) |
| 2θ range (°) | 3.469–31.506 |
| Reflections collected | 70043 |
| Independent reflections | 23878 |
| Goodness–of–fit (GOF) | 1.073 |
| R [I ≥ 2σ (I)] | 0.0539 |
| wR2 [I ≥ 2σ (I)] | 0.1289 |
| D–H…A | D–H | H…A | D…A | D–H…A |
|---|---|---|---|---|
| N1A–H1A…O1A | 0.80 (4) | 2.00 (4) | 2.642 (3) | 137 (3) |
| N1B–H1B…O1B | 0.84 (3) | 2.03 (3) | 2.686 (3) | 134 (3) |
| N1C–H1C…O2C | 0.88 (3) | 1.93 (3) | 2.643 (3) | 138 (3) |
| N1D–H1D…O1D | 0.83 (3) | 2.02 (3) | 2.671 (3) | 134 (3) |
| N1E–H1E…O1E | 0.86 (3) | 2.10 (3) | 2.718 (3) | 129 (3) |
| N1F–H1F…O1F | 0.88 (3) | 1.92 (4) | 2.638 (3) | 138 (4) |
| O1G–H1G…O1I | 0.68 (3) | 1.99 (3) | 2.636 (3) | 160 (4) |
| O1I–H1I…O1KA | 0.74 (5) | 2.02 (5) | 2.751 (4) | 170 (4) |
| O1KA–H1KA…O1L | 0.82 | 1.85 | 2.665 (5) | 173 |
| O1L–H1L…O1KAa | 0.82 | 2.03 | 2.824 (5) | 163 |
| Chemicals Microorganism | 1 | 2 | 3 | 4 | 5 | 6 | Van/Nys |
|---|---|---|---|---|---|---|---|
| MIC [mg/L] | |||||||
| Gram-Positive bacteria | Van | ||||||
| S. aureus ATCC 25923 | 62.5 | 62.5 | 62.5 | 62.5 | 62.5 | 62.5 | 0.98 |
| S. epidermidis ATCC 12228 | 62.5 | 62.5 | 62.5 | 62.5 | 62.5 | 62.5 | 0.98 |
| M. luteus ATCC 10240 | 125 | 62.5 | 62.5 | 62.5 | 31.3 | 62.5 | 0.12 |
| E. faecalis ATCC 29212 | 500 | 500 | 500 | 500 | 500 | 500 | 1.95 |
| B. subtilis ATCC 6633 | 62.5 | 62.5 | 62.5 | 62.5 | 62.5 | 62.5 | 0.24 |
| B. cereus ATCC 10876 | 62.5 | 31.3 | 62.5 | 62.5 | 62.5 | 31.3 | 0.98 |
| S. pyogenes ATCC 19615 | 125 | 125 | 125 | 125 | 125 | 125 | 0.24 |
| S. pneumoniae ATCC 49619 | 62.5 | 62.5 | 125 | 125 | 125 | 125 | 0.24 |
| S. mutans ATCC 25175 | 250 | 250 | 250 | 250 | 250 | 125 | 0.98 |
| Gram-Negative bacteria | Cip | ||||||
| S. typhimurium ATCC 14028 | >1000 | >1000 | >1000 | >1000 | >1000 | >1000 | 0.061 |
| E. coli ATCC 25922 | >1000 | >1000 | >1000 | >1000 | >1000 | >1000 | 0.015 |
| P. mirabilis ATCC 12453 | 250 | 250 | 250 | 250 | 250 | 250 | 0.030 |
| K. pneumoniae ATCC 13883 | >1000 | >1000 | >1000 | >1000 | >1000 | >1000 | 0.122 |
| P. aeruginosa ATCC 9027 | >1000 | >1000 | >1000 | >1000 | >1000 | >1000 | 0.488 |
| Yeasts | Nys | ||||||
| C. albicans ATCC 2091 | 250 | 250 | 250 | 250 | 250 | 250 | 0.48 |
| C. parapsilosis ATCC 22019 | 250 | 250 | 250 | 250 | 250 | 250 | 0.24 |
| C. glabrata ATCC 90030 | 500 | 250 | 500 | 500 | 500 | 500 | 0.24 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gacki, M.; Kafarska, K.; Pietrzak, A.; Szczesio, M.; Korona-Głowniak, I.; Wolf, W.M. Transition Metal Complexes with Flufenamic Acid for Pharmaceutical Applications—A Novel Three-Centered Coordination Polymer of Mn(II) Flufenamate. Materials 2020, 13, 3705. https://doi.org/10.3390/ma13173705
Gacki M, Kafarska K, Pietrzak A, Szczesio M, Korona-Głowniak I, Wolf WM. Transition Metal Complexes with Flufenamic Acid for Pharmaceutical Applications—A Novel Three-Centered Coordination Polymer of Mn(II) Flufenamate. Materials. 2020; 13(17):3705. https://doi.org/10.3390/ma13173705
Chicago/Turabian StyleGacki, Michał, Karolina Kafarska, Anna Pietrzak, Małgorzata Szczesio, Izabela Korona-Głowniak, and Wojciech M. Wolf. 2020. "Transition Metal Complexes with Flufenamic Acid for Pharmaceutical Applications—A Novel Three-Centered Coordination Polymer of Mn(II) Flufenamate" Materials 13, no. 17: 3705. https://doi.org/10.3390/ma13173705
APA StyleGacki, M., Kafarska, K., Pietrzak, A., Szczesio, M., Korona-Głowniak, I., & Wolf, W. M. (2020). Transition Metal Complexes with Flufenamic Acid for Pharmaceutical Applications—A Novel Three-Centered Coordination Polymer of Mn(II) Flufenamate. Materials, 13(17), 3705. https://doi.org/10.3390/ma13173705