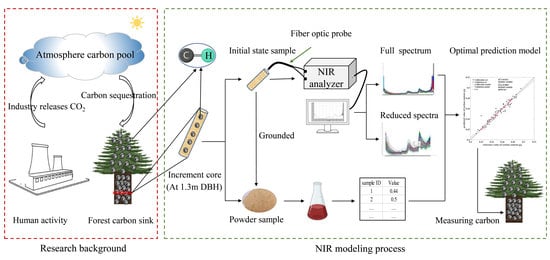
Prediction of the Carbon Content of Six Tree Species from Visible-Near-Infrared Spectroscopy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Carbon Content Based on Chemical Analysis
2.3. Spectra Collection
2.4. Pre-Processing of Spectroscopic Data
2.5. Model Development
2.6. Model Evaluation
2.7. Software
3. Results and Discussion
3.1. Near-Infrared Spectral Features
3.2. The Selection of Sample Sets
3.3. PLS Model Development
3.4. Reduced Spectra Model
3.5. Comparison of Carbon Content of Tree Species
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Al-Ghussain, L. Global warming: Review on driving forces and mitigation. Environ. Prog. Sustain. Energy 2019, 38, 13–21. [Google Scholar] [CrossRef] [Green Version]
- Scheffer, M.; Brovkin, V.; Cox, P.M. Positive feedback between global warming and atmospheric CO2 concentration inferred from past climate change. Geophys. Res. Lett. 2006, 33. [Google Scholar] [CrossRef]
- Wang, J.; Quan, Q.; Chen, W.; Tian, D.; Ciais, P.; Crowther, T.W.; Mack, M.C.; Poulter, B.; Tian, H.; Luo, Y.; et al. Increased CO2 emissions surpass reductions of non-CO2 emissions more under higher experimental warming in an alpine meadow. Sci. Total Environ. 2021, 769, 144559. [Google Scholar] [CrossRef]
- Węgiel, A.; Polowy, K. Aboveground Carbon Content and Storage in Mature Scots Pine Stands of Different Densities. Forests 2020, 11, 240. [Google Scholar] [CrossRef] [Green Version]
- Carvalhais, N.; Forkel, M.; Khomik, M.; Bellarby, J.; Jung, M.; Migliavacca, M.; Mu, M.; Saatchi, S.; Santoro, M.; Thurner, M.; et al. Global covariation of carbon turnover times with climate in terrestrial ecosystems. Nature 2014, 514, 213–217. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.Y.; Duan, A.G. Advances in Carbon Reserve of Forest Ecosystems. For. Sci. Technol. 2020, 2, 3–6. [Google Scholar] [CrossRef]
- Vergara-Díaz, G.; Herrera-Machuca, M.Á. Estimation and spatial analysis of aerial biomass and carbon capture in native forests in the south of Chile: County of Valdivia. Rev. Chapingo Ser. Cienc. For. Ambiente 2020, 27, 53–71. [Google Scholar] [CrossRef]
- Solomon, N.; Birhane, E.; Tadesse, T.; Treydte, A.C.; Meles, K. Carbon stocks and sequestration potential of dry forests under community management in Tigray, Ethiopia. Ecol. Process. 2017, 6, 20. [Google Scholar] [CrossRef]
- Lu, J.; Feng, Z.; Zhu, Y. Estimation of Forest Biomass and Carbon Storage in China Based on Forest Resources Inventory Data. Forests 2019, 10, 650. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Fan, W.Y.; Li, M.Z. Forest carbon rates at different scales in Northeast China forest area. Chin. J. Appl. Ecol. 2012, 23, 341–346. [Google Scholar] [CrossRef]
- Li, B.; Fang, X.; Tian, D.L. Studies on carbon concentration of main forest vegetation tree species in Hunan province. J. Cent. South Univ. For. Technol. 2015, 1, 71–78. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, C.; Wang, X.; Quan, X. Carbon concentration variability of 10 Chinese temperate tree species. For. Ecol. Manag. 2009, 258, 722–727. [Google Scholar] [CrossRef]
- Dong, L.; Liu, Y.; Zhang, L.; Xie, L.; Li, F. Variation in Carbon Concentration and Allometric Equations for Estimating Tree Carbon Contents of 10 Broadleaf Species in Natural Forests in Northeast China. Forests 2019, 10, 928. [Google Scholar] [CrossRef] [Green Version]
- Pompa-García, M.; Sigala-Rodríguez, J.A.; Jurado, E.; Flores, J. Tissue carbon concentration of 175 Mexican forest species. Iforest—Biogeosci. For. 2017, 10, 754–758. [Google Scholar] [CrossRef] [Green Version]
- Gómez-García, E. Estimating the changes in tree carbon stocks in Galician forests (NW Spain) between 1972 and 2009. For. Ecol. Manag. 2020, 467, 118157. [Google Scholar] [CrossRef]
- Xu, Q.; Lin, L.; Xue, C.; Luo, Y.; Lei, Y. Component specific carbon content and storage of Cinnamomum camphora in Guangdong Province. J. Zhejiang AF Univ. 2019, 36, 70–79. [Google Scholar] [CrossRef]
- Sun, X.; Li, H.; Yi, Y.; Hua, H.; Guan, Y.; Chen, C. Rapid detection and quantification of adulteration in Chinese hawthorn fruits powder by near-infrared spectroscopy combined with chemometrics. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 250, 119346. [Google Scholar] [CrossRef]
- Li, J.Y.; Chu, X.L.; Chen, P.; Tian, S.B. Application of Spectral Automatic Retrieval Algorithm on the Rapid Establishment of Gasoline Spectral Database. Acta Pet. Sin. 2017, 1, 131–137. [Google Scholar] [CrossRef]
- He, K.; Zhong, M.; Li, Z.; Liu, J. Near-infrared spectroscopy for the concurrent quality prediction and status monitoring of gasoline blending. Control. Eng. Pract. 2020, 101, 104478. [Google Scholar] [CrossRef]
- Mishra, P.; Herrmann, I.; Angileri, M. Improved prediction of potassium and nitrogen in dried bell pepper leaves with visible and near-infrared spectroscopy utilising wavelength selection techniques. Talanta 2021, 225, 121971. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.D.; Wang, Y.B.; Wang, R.J.; Wang, L.S.; Lu, C.P.; Zhang, Z.Y.; Song, L.T.; Liu, Y. Improvements of the Vis-NIRS Model in the Prediction of Soil Organic Matter Content Using Spectral Pretreatments, Sample Selection, and Wavelength Optimization. J. Appl. Spectrosc. 2017, 84, 529–534. [Google Scholar] [CrossRef]
- Shao, Y.; Jiang, L.; Zhou, H.; Pan, J.; He, Y. Identification of pesticide varieties by testing microalgae using Visible/Near Infrared Hyperspectral Imaging technology. Sci. Rep. 2016, 6, 24221. [Google Scholar] [CrossRef] [Green Version]
- Santos, I.A.; Conceicao, D.G.; Viana, M.B.; Silva, G.J.; Santos, L.S.; Ferrao, S.P.B. NIR and MIR spectroscopy for quick detection of the adulteration of cocoa content in chocolates. Food Chem. 2021, 349, 129095. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Li, C.; Takeda, F. Nondestructive Detection and Quantification of Blueberry Bruising using Near-infrared (NIR) Hyperspectral Reflectance Imaging. Sci. Rep. 2016, 6, 35679. [Google Scholar] [CrossRef] [Green Version]
- Lakeh, M.A.; Karimvand, S.K.; Khoshayand, M.R.; Abdollahi, H. Analysis of residual moisture in a freeze-dried sample drug using a multivariate fitting regression model. Microchem. J. 2020, 154, 104516. [Google Scholar] [CrossRef]
- Ma, Y.; He, H.; Wu, J.; Wang, C.; Chao, K.; Huang, Q. Assessment of Polysaccharides from Mycelia of genus Ganoderma by Mid-Infrared and Near-Infrared Spectroscopy. Sci. Rep. 2018, 8, 10. [Google Scholar] [CrossRef] [PubMed]
- Hobley, E.; Steffens, M.; Bauke, S.L.; Kogel-Knabner, I. Hotspots of soil organic carbon storage revealed by laboratory hyperspectral imaging. Sci. Rep. 2018, 8, 13900. [Google Scholar] [CrossRef]
- Blaschek, M.; Roudier, P.; Poggio, M.; Hedley, C.B. Prediction of soil available water-holding capacity from visible near-infrared reflectance spectra. Sci. Rep. 2019, 9, 12833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amaral, E.A.; Dos Santos, L.M.; Hein, P.R.G.; Costa, E.V.S.; Rosado, S.C.S.; Trugilho, P.F. Evaluating basic density calibrations based on NIR spectra recorded on the three wood faces and subject to different mathematical treatments. N. Z. J. For. Sci. 2021, 51. [Google Scholar] [CrossRef]
- Pace, J.-H.C.; Latorraca, J.-V.D.F.; Hein, P.-R.G.; Carvalho, A.M.d.; Castro, J.P.; Silva, C.-E.S.d. Wood species identification from Atlantic forest by near infrared spectroscopy. For. Syst. 2019, 28, e015. [Google Scholar] [CrossRef]
- Li, Y.; Via, B.K.; Young, T.; Li, Y. Visible-Near Infrared Spectroscopy and Chemometric Methods for Wood Density Prediction and Origin/Species Identification. Forests 2019, 10, 1078. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Via, B.K.; Cheng, Q. New Pretreatment Methods for Visible–Near-Infrared Calibration Modeling of Air-Dry Density of Ulmus pumila Wood. For. Prod. J. 2019, 69, 188–194. [Google Scholar] [CrossRef]
- Herrmann, S.; Bauhus, J. Nutrient retention and release in coarse woody debris of three important central European tree species and the use of NIRS to determine deadwood chemical properties. For. Ecosyst. 2018, 5, 22. [Google Scholar] [CrossRef] [Green Version]
- Elle, O.; Richter, R.; Vohland, M.; Weigelt, A. Fine root lignin content is well predictable with near-infrared spectroscopy. Sci. Rep. 2019, 9, 6396. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sun, C.; Zhou, B.; He, Y. Determination of Hemicellulose, Cellulose and Lignin in Moso Bamboo by Near Infrared Spectroscopy. Sci. Rep. 2015, 5, 17210. [Google Scholar] [CrossRef] [PubMed]
- Murguzur, F.J.A.; Bison, M.; Smis, A.; Bohner, H.; Struyf, E.; Meire, P.; Brathen, K.A. Towards a global arctic-alpine model for Near-infrared reflectance spectroscopy (NIRS) predictions of foliar nitrogen, phosphorus and carbon content. Sci. Rep. 2019, 9, 8259. [Google Scholar] [CrossRef] [PubMed]
- Hou, R.; Ji, H.Y.; Zhang, D.L. Quantitative analysis of barley protein content based on OSC PLS algorithm. Spectrosc. Spectr. Anal. 2009, 7, 1840–1843. [Google Scholar] [CrossRef]
- Mareclo, A.M.; Cristiano, G.F. Determination of protection in field cru by specictroscopy NIR and PLS1. Sci. Technol. Food. Camp. 2005, 25, 25–31. [Google Scholar] [CrossRef] [Green Version]
- Kawamura, K.; Nishigaki, T.; Tsujimoto, Y.; Andriamananjara, A.; Rabenaribo, M.; Asai, H.; Rakotoson, T.; Razafimbelo, T. Exploring relevant wavelength regions for estimating soil total carbon contents of rice fields in Madagascar from Vis-NIR spectra with sequential application of backward interval PLS. Plant Prod. Sci. 2020, 24, 1–14. [Google Scholar] [CrossRef]
- Kuligowski, J.; Carrión, D.; Quintás, G.; Garrigues, S.; de la Guardia, M. Direct determination of polymerised triacylglycerides in deep-frying vegetable oil by near infrared spectroscopy using Partial Least Squares regression. Food Chem. 2012, 131, 353–359. [Google Scholar] [CrossRef]
- Pereira, H.; Santos, A.J.A.; Anjos, O. Fibre Morphological Characteristics of Kraft Pulps of Acacia melanoxylon Estimated by NIR-PLS-R Models. Materials 2016, 9, 8. [Google Scholar] [CrossRef] [Green Version]
- Costa, M.C.A.; Morgano, M.A.; Ferreira, M.M.C.; Milani, R.F. Analysis of bee pollen constituents from different Brazilian regions: Quantification by NIR spectroscopy and PLS regression. LWT 2017, 80, 76–83. [Google Scholar] [CrossRef]
- Pereira, E.V.d.S.; Fernandes, D.D.d.S.; de Araújo, M.C.U.; Diniz, P.H.G.D.; Maciel, M.I.S. Simultaneous determination of goat milk adulteration with cow milk and their fat and protein contents using NIR spectroscopy and PLS algorithms. LWT 2020, 127, 109427. [Google Scholar] [CrossRef]
- Biney, J.K.M.; Blöcher, J.R.; Borůvka, L.; Vašát, R. Does the limited use of orthogonal signal correction pre-treatment approach to improve the prediction accuracy of soil organic carbon need attention? Geoderma 2021, 388, 114945. [Google Scholar] [CrossRef]
- Yin, S.K.; Li, C.X.; Meng, Y.B. Near infrared spectral estimation and model optimization of Tilia tuan based on different pretreatments. J. Cent. South Univ. For. Technol. 2020, 40, 176–185. [Google Scholar] [CrossRef]
- Lu, B.; Wang, X.; Liu, N.; Hu, C.; Xu, H.; Wu, K.; Xiong, Z.; Tang, X. Quantitative NIR spectroscopy determination of coco-peat substrate moisture content: Effect of particle size and non-uniformity. Infrared Phys. Technol. 2020, 111, 103482. [Google Scholar] [CrossRef]
- Hao, Y.; Geng, P.; Wu, W.; Wen, Q.; Rao, M. Identification of Rice Varieties and Transgenic Characteristics Based on Near-Infrared Diffuse Reflectance Spectroscopy and Chemometrics. Molecules 2019, 24, 4568. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Liu, Y.; Chen, Y.; Zhang, Y.; Shi, T.; Wang, J.; Hong, Y.; Fei, T. The Influence of Spectral Pretreatment on the Selection of Representative Calibration Samples for Soil Organic Matter Estimation Using Vis-NIR Reflectance Spectroscopy. Remote Sens. 2019, 11, 450. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.M.; Ji, J.M.; Gao, H.Z. The Effect of MSC Spectral Pretreatment Regions on Near Infrared Spectroscopy Calibration Results. Spectrosc. Spectr. Anal. 2014, 34, 2387–2390. [Google Scholar] [CrossRef]
- Taylor, A.R.; Wang, J.R.; Kurz, W.A. Effects of harvesting intensity on carbon stocks in eastern Canadian red spruce (Picea rubens) forests: An exploratory analysis using the CBM-CFS3 simulation model. For. Ecol. Manag. 2008, 255, 3632–3641. [Google Scholar] [CrossRef]
- Jiang, Z.H.; Huang, A.M.; Wang, B. Near Infrared Spectroscopy of Wood Sections and Rapid Density Prediction. Spectrosc. Spectr. Anal. 2006, 26, 1034–1037. [Google Scholar] [CrossRef]
- Zhu, H.; Chu, B.; Fan, Y.; Tao, X.; Yin, W.; He, Y. Hyperspectral Imaging for Predicting the Internal Quality of Kiwifruits Based on Variable Selection Algorithms and Chemometric Models. Sci. Rep. 2017, 7, 7845. [Google Scholar] [CrossRef] [Green Version]
- Lequeue, G.; Draye, X.; Baeten, V. Determination by near infrared microscopy of the nitrogen and carbon content of tomato (Solanum lycopersicum L.) leaf powder. Sci. Rep. 2016, 6, 33183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krahmer, A.; Engel, A.; Kadow, D.; Ali, N.; Umaharan, P.; Kroh, L.W.; Schulz, H. Fast and neat--determination of biochemical quality parameters in cocoa using near infrared spectroscopy. Food Chem. 2015, 181, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Vergnoux, A.; Dupuy, N.; Guiliano, M.; Vennetier, M.; Theraulaz, F.; Doumenq, P. Fire impact on forest soils evaluated using near-infrared spectroscopy and multivariate calibration. Talanta 2009, 80, 39–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhan, X.R.; Zhu, X.R.; Shi, X.Y. Determination of Hesperidin in Tangerine Leaf by Near-Infrared Spectroscopy with SPXY Algorithm for Sample Subset Partitioning and Monte Carlo Cross Validation. Spectrosc. Spectr. Anal. 2009, 29, 964–968. [Google Scholar] [CrossRef]
- Wang, S.F.; Han, P.; Cui, G.L.; Wang, D.; Liu, S.S.; Zhao, Y. The NIR Detection Research of Soluble Solid Content in Watermelon Based on SPXY Algorithm. Spectrosc. Spectr. Anal. 2019, 39, 738–742. [Google Scholar] [CrossRef]
- Rinnan, Å.; van den Berg, F.; Engelsen, S.B. Review of the most common pre-processing techniques for near-infrared spectra. TrAC Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Liu, L.; Ye, X.P.; Saxton, A.M.; Womac, A. Pretreatment of near Infrared Spectral Data in Fast Biomass Analysis. J. Near Infrared Spectrosc. 2010, 18, 317–331. [Google Scholar] [CrossRef]
- Zhao, L.Z.; Guo, Y.; Dou, Y.; Wang, B.; Mi, H.; Ren, Y.L. Application of artificial neural networks to the nondestructive determination of ciprofloxacin hydrochloride in powder by short-wavelength NIR spectroscopy. J. Anal. Chem. 2007, 62, 1156–1162. [Google Scholar] [CrossRef]
- Xiong, Y.; Ohashi, S.; Nakano, K.; Jiang, W.; Takizawa, K.; Iijima, K.; Maniwara, P. Application of the radial basis function neural networks to improve the nondestructive Vis/NIR spectrophotometric analysis of potassium in fresh lettuces. J. Food Eng. 2021, 298, 110417. [Google Scholar] [CrossRef]
- Tian, W.; Chen, G.; Zhang, G.; Wang, D.; Tilley, M.; Li, Y. Rapid determination of total phenolic content of whole wheat flour using near-infrared spectroscopy and chemometrics. Food Chem. 2021, 344, 128633. [Google Scholar] [CrossRef]
- Faber, N.M.; Rajko, R. How to avoid over-fitting in multivariate calibration--the conventional validation approach and an alternative. Anal. Chim. Acta 2007, 595, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Hadoux, X.; Gorretta, N.; Roger, J.-M.; Bendoula, R.; Rabatel, G. Comparison of the efficacy of spectral pre-treatments for wheat and weed discrimination in outdoor conditions. Comput. Electron. Agric. 2014, 108, 242–249. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.S.; Qi, D.W.; Huang, A.M. Denoising of Near Infrared Spectroscopy in Wood Based on Wavelet TransformModulus Maximum. Sci. Silvae Sin. 2008, 10, 109–112. [Google Scholar] [CrossRef]
- Barbin, D.F.; Felicio, A.L.d.S.M.; Sun, D.-W.; Nixdorf, S.L.; Hirooka, E.Y. Application of infrared spectral techniques on quality and compositional attributes of coffee: An overview. Food Res. Int. 2014, 61, 23–32. [Google Scholar] [CrossRef] [Green Version]
- Westad, F.; Schmidt, A.; Kermit, M. Incorporating Chemical Band-Assignment in near Infrared Spectroscopy Regression Models. J. Near Infrared Spectrosc. 2008, 16, 265–273. [Google Scholar] [CrossRef]







| Tree Species Name | Number of Samples | Minimum (g/g) | Maximum (g/g) | Average Value (g/g) | Standard Deviation (g/g) |
|---|---|---|---|---|---|
| Betula platyphylla | 15 | 0.4200 | 0.4590 | 0.4366 | 0.0106 |
| Abies fabri (Mast.) Craib | 15 | 0.4500 | 0.4950 | 0.4752 | 0.0126 |
| Larix gmelinii | 15 | 0.4320 | 0.4800 | 0.4564 | 0.0155 |
| Acer tegmentosum Maxim. | 15 | 0.4110 | 0.4740 | 0.4364 | 0.0154 |
| Acer pictum Thunb. ex Murray | 16 | 0.4110 | 0.4710 | 0.4397 | 0.0178 |
| Picea asperata Mast. | 20 | 0.4400 | 0.4780 | 0.4622 | 0.0104 |
| total | 96 | 0.4110 | 0.4950 | 0.4515 | 0.0198 |
| Sample Set Name | Number of Samples | Minimum (g/g) | Maximum (g/g) | Average Value (g/g) | Standard Deviation (g/g) | Coefficient of Variation (%) |
|---|---|---|---|---|---|---|
| Calibration set | 64 | 0.4110 | 0.4952 | 0.4533 | 0.0203 | 4.48 |
| Validation set | 32 | 0.4200 | 0.4770 | 0.4486 | 0.0177 | 3.95 |
| Spectral Pretreatment Method | Cross-Validation | Calibration Set | Validation Set | ||||
|---|---|---|---|---|---|---|---|
| OPC | RMSECV | R2c | RMSEC | R2p | RMSEP | RPD | |
| Raw | 2 | 0.0154 | 0.65 | 0.0120 | 0.72 | 0.0091 | 1.9 |
| EMSC | 4 | 0.0136 | 0.80 | 0.0090 | 0.95 | 0.0037 | 4.8 |
| 1D | 8 | 0.0186 | 0.92 | 0.0056 | 0.99 | 0.0020 | 8.9 |
| 2D | 10 | 0.0203 | 0.91 | 0.0059 | 0.98 | 0.0023 | 7.7 |
| Baseline correction | 4 | 0.0158 | 0.73 | 0.0104 | 0.79 | 0.0081 | 2.2 |
| de-trend | 3 | 0.0173 | 0.77 | 0.0096 | 0.86 | 0.0064 | 2.8 |
| OSC | 3 | 0.0136 | 0.81 | 0.0088 | 0.95 | 0.0037 | 4.8 |
| normalization | 5 | 0.0156 | 0.88 | 0.0071 | 0.96 | 0.0034 | 5.2 |
| Spectra and Wavelength | Spectral Pretreatment Method | Cross-Validation | Calibration Set | Validation Set | |||
|---|---|---|---|---|---|---|---|
| OPC | RMSECV | R2c | RMSEC | R2p | RMSEP | ||
| Full Spectra 350–2500 nm | Raw | 2 | 0.0154 | 0.6467 | 0.0120 | 0.7242 | 0.0091 |
| Reduced spectra 400–2350 nm | Raw | 7 | 0.0147 | 0.69 | 0.0111 | 0.87 | 0.0064 |
| EMSC | 2 | 0.0161 | 0.46 | 0.0148 | 0.63 | 0.0105 | |
| 1D | 2 | 0.0150 | 0.53 | 0.0139 | 0.70 | 0.0095 | |
| 2D | 2 | 0.0150 | 0.59 | 0.0129 | 0.73 | 0.0091 | |
| Baseline correction | 3 | 0.0147 | 0.53 | 0.0138 | 0.65 | 0.0102 | |
| de-trend | 5 | 0.0157 | 0.64 | 0.0121 | 0.86 | 0.0065 | |
| OSC | 6 | 0.0142 | 0.69 | 0.0112 | 0.99 | 0.0001 | |
| normalization | 1 | 0.0148 | 0.50 | 0.0142 | 0.63 | 0.0109 | |
| Tree Species Name | Number of Samples | Minimum (g/g) | Maximum (g/g) | Average Valu (g/g) | Standard Deviation (g/g) |
|---|---|---|---|---|---|
| Betula platyphylla | 15 | 0.4196 | 0.4598 | 0.4360 d | 0.0110 |
| Abies fabri (Mast.) Craib | 15 | 0.4517 | 0.4985 | 0.4745 a | 0.0121 |
| Larix gmelinii | 15 | 0.4369 | 0.4836 | 0.4559 bc | 0.0146 |
| Acer tegmentosum Maxim. | 15 | 0.4117 | 0.4684 | 0.4352 d | 0.0148 |
| Acer pictum Thunb. ex Murray | 16 | 0.4176 | 0.4763 | 0.4430 cd | 0.0144 |
| Picea asperata Mast. | 20 | 0.4435 | 0.4788 | 0.4627 ab | 0.0095 |
| total | 96 | 0.4117 | 0.4983 | 0.4517 | 0.0189 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, Y.; Zhang, Y.; Li, C.; Zhao, J.; Wang, Z.; Wang, C.; Li, Y. Prediction of the Carbon Content of Six Tree Species from Visible-Near-Infrared Spectroscopy. Forests 2021, 12, 1233. https://doi.org/10.3390/f12091233
Meng Y, Zhang Y, Li C, Zhao J, Wang Z, Wang C, Li Y. Prediction of the Carbon Content of Six Tree Species from Visible-Near-Infrared Spectroscopy. Forests. 2021; 12(9):1233. https://doi.org/10.3390/f12091233
Chicago/Turabian StyleMeng, Yongbin, Yuanyuan Zhang, Chunxu Li, Jinghan Zhao, Zichun Wang, Chen Wang, and Yaoxiang Li. 2021. "Prediction of the Carbon Content of Six Tree Species from Visible-Near-Infrared Spectroscopy" Forests 12, no. 9: 1233. https://doi.org/10.3390/f12091233
APA StyleMeng, Y., Zhang, Y., Li, C., Zhao, J., Wang, Z., Wang, C., & Li, Y. (2021). Prediction of the Carbon Content of Six Tree Species from Visible-Near-Infrared Spectroscopy. Forests, 12(9), 1233. https://doi.org/10.3390/f12091233