Former Land Use and Host Genotype Influence the Mycorrhizal Colonization of Poplar Roots
Abstract
:1. Introduction
2. Experimental Section
2.1. Plant Material, Study Site and Sample Collection
2.2. Ectomycorrhizas (ECM)
2.3. Arbuscular Mycorrhiza (AM)
2.4. Molecular Identification of ECM
2.5. Molecular Detection of AM
2.6. Statistical Analyses
3. Results and Discussion
3.1. Richness and Diversity of Mycorrhiza

| Soil and Clone | Species Richness | Shannon’s Diversity Index | Evenness Index |
|---|---|---|---|
| S | 12 | 3.15 | 1.27 |
| P | 8 | 3.24 | 1.56 |
| G | 4 | 1.83 | 1.32 |
| C | 3 | 2.26 | 2.06 |
| C1 | 10 | 2.37 | 1.03 |
| C2 | 8 | 1.87 | 0.90 |
| C3 | 6 | 2.18 | 1.22 |
| C4 | 8 | 2.31 | 1.11 |
| C5 | 8 | 2.07 | 1.00 |
| C6 | 8 | 2.35 | 1.13 |
| C7 | 8 | 2.19 | 1.05 |
| Mycorrhizal Taxon | GenBank Accesion Number |
|---|---|
| Inocybe curvipes | KJ591031, KJ591036, KJ591039 KJ591030, KJ591037, KJ591038 |
| Paxillus involutus | KJ591032 |
| Hebeloma sacchariolens | KJ624710 |
| Hebeloma vaccinum | KJ591035 |
| Laccaria tortilis | KJ591042, KJ591043, KJ591044 |
| Laccaria proxima | KJ591040, KJ591041 |
| Laccaria bicolor | KJ624712 |
| Pezizales (1) | KJ591033 |
| Pezizaceae (2) | KJ591045 |
| Pezizales (3) | KJ591034 |
| Pezizales (4) | KJ624711 |
| Pezizales (5) | KJ624714 |
| Sebacinales | KJ624715 |
| Meliniomyces | KJ624713 |
| Glomus mossae | KJ591046 |
| Glomus intraradices | KJ591047 |
| Glomus hoi | KJ591049 |
| Scutellospora sp. | KJ591048 |
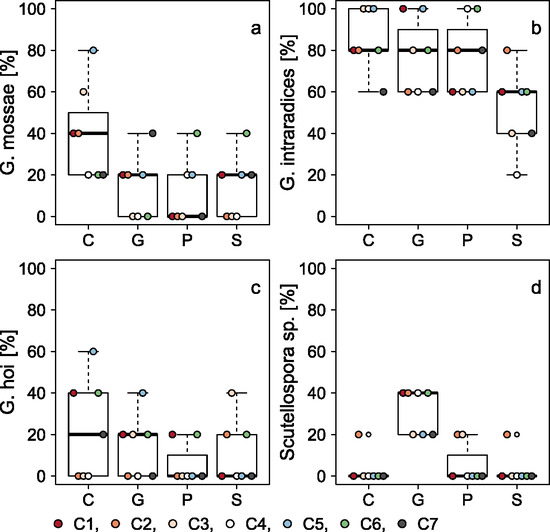
3.2. Influence of the Former Land Use Type
| Response Variable | Transformation | F-Value | p-Value |
|---|---|---|---|
| AM | - | 8.05 | 0.0013018 |
| ECM | - | 0.85 | 0.4867 |
| Shoot length | - | 55.86 | <0.001 |
| AM hyphae | squared | 9.77 | <0.001 |
| Vesicles | rank a | 7.20 | 0.002245 |
| Arbuscules | rank a | 0.58 | 0.634039 |
| Glomus mossae | - | 3.16 | 0.04999 |
| Glomus intraradices | - | 4.36 | 0.01787 |
| Glomus hoi | - | 1.45 | 0.261 |
| Scutellospora sp. | - | 28.3 | <0.001 |
| Paxillus involutus | rank a | 36.67 | <0.001 |
| Laccaria tortilis | log10 + 1 | 6.31 | 0.0041 |
| L. proxima | rank a | 6.99 | 0.00258 |
| Former Land Use Type | C | N | P | S | pH (CaCl2) | pH (H2O) |
|---|---|---|---|---|---|---|
| Cornfield | 12.94 | 1.32 | 0.67 | 0.19 | 5.07 | 5.81 |
| Grassland | 10.13 | 0.95 | 0.58 | 0.13 | 5.59 | 6.23 |
| Poplar stand | 18.72 | 1.74 | 0.58 | 0.24 | 5.49 | 6.11 |
| Spruce forest | 47.70 | 2.62 | 0.36 | 0.34 | 3.50 | 4.12 |




3.3. Influence of the Clone Type
| Response Variable | Transformation | F-Value | p-Value |
|---|---|---|---|
| AM | - | 8.97 | <0.001 |
| ECM | - | 1.47 | 0.2437 |
| Shoot length | - | 32.35 | <0.001 |
| AM hyphae | - | 1.55 | 0.218383 |
| Vesicles | - | 18.93 | <0.001 |
| Arbuscules | log10+1 | 14.85 | <0.001 |
| Glomus mossae | log10+1 | 1.10 | 0.40137 |
| Glomus intraradices | - | 0.55 | 0.76256 |
| Glomus hoi | log10+1 | 2.04 | 0.1132 |
| Scutellospora sp. | log10+1 | 5.08 | 0.003283 |
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References and Notes
- Langer, J.; Santner, D.; Krpata, W.J.; Fitz, W.W.; Wenzel, P.; Schweiger, F. Ectomycorrhizal impact on Zn accumulation of Populus tremula L. grown in metalliferous soil with increasing levels of Zn concentration. Plant Soil 2012, 355, 283–297. [Google Scholar] [CrossRef]
- Bradshaw, H.D.; Reinhart, C.; Davis, J.; Stettler, R. Emerging model systems in plant biology: Poplar (Populus) as a model forest tree. J. Plant Growth Regul. 2000, 19, 306–313. [Google Scholar] [CrossRef]
- Brundrett, M.C. Mycorrhizal associations and other means of nutrition of vascular plants: Understanding the global diversity of host plants by resolving conflicting information and developing reliable means of diagnosis. Plant Soil 2009, 320, 37–77. [Google Scholar] [CrossRef]
- Pullman, G.S.; Cairney, J.; Peter, G. Clonal forestry and genetic engineering: Forest biotechnology-where we stand and future prospects and impacts. Tappi J. 1998, 81, 57–64. [Google Scholar]
- Tedersoo, L.; May, T.W.; Smith, M.E. Ectomycorrhizal lifestyle in fungi: Global diversity, distribution, and evolution of phylogenetic lineages. Mycorrhiza 2010, 20, 217–263. [Google Scholar] [CrossRef] [PubMed]
- Lodge, D.J. The influence of soil moisture and flooding on formation of VA-endo and ectomycorrhizae in Populus and Salix. Plant Soil 1989, 117, 243–253. [Google Scholar] [CrossRef]
- Molina, R.; Massicotte, H.; Trappe, J.M. Specificity phenomena in mycorrhizal symbioses: Community-ecological consequences and practical implications. In Routledge AMF ycorrhizal Functioning, an Integrative Plant—Fungal Process; Chapman & Hall, Inc.: New York, NY, USA, 1992; pp. 357–423. [Google Scholar]
- Gehring, C.A.; Mueller, C.; Whitham, T.G. Environmental and genetic effects on the formation of ectomycorrhizal and arbuscular mycorrhizal associations in cottonwoods. Oecologia 2006, 149, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.; Read, D. Mycorrhizal Symbiosis, 2nd ed.; Academic Press: London, UK, 1997. [Google Scholar]
- Khasa, P.D.; Chakravarty, P.; Robertson, A.; Thomas, B.R.; Dancik, B.P. The mycorrhizal status of selected poplar clones introduced in Alberta. Biomass Bioenergy 2002, 22, 99–104. [Google Scholar] [CrossRef]
- Tagu, D.; Faivre-Rampant, P.; Lapeyrie, F.; Frey-Klett, P.; Vion, P.; Villar, M. Variation in the ability to form ectomycorrhizas in the F1 progeny of an interpsecic poplar Populus spp. Cross. Mycorrhiza 2001, 10, 237–240. [Google Scholar] [CrossRef]
- Tagu, D.; Bastien, C.; Faivre-Rampant, P.; Garbaye, J.; Vion, P.; Martin, F. Genetic analysis of phenotypic variation for ectomycorrhiza formation in an interspecic F1 poplar full-sib family. Mycorrhiza 2005, 15, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Van der Heijden, E.W.; Kuyper, T.W. Laboratory experiments imply the conditionality of mycorrhizal benefits for Salix repens: Role of pH and nitrogen to phosphorus ratios. Plant Soil 2001, 228, 275–290. [Google Scholar] [CrossRef]
- Walker, C.; McNabb, H.S. Mycorrhizal symbionts associated with hybrid poplars from Iowa, USA. Eur. J. Path 1984, 14, 282–296. [Google Scholar] [CrossRef]
- Neville, J.; Tessier, J.L.; Morrison, I.; Scarratt, J.; Canning, B.; Klironomos, J.N. Soil depth distribution of ecto- and arbuscular mycorrhizal fungi associated with Populus tremuloides within a 3-year-old boreal forest clear-cut. Appl. Soil Ecol. 2002, 19, 209–216. [Google Scholar] [CrossRef]
- Danielsen, L.; Thürmer, A.; Meinicke, P.; Buée, M.; Morin, E.; Martin, F.; Pilate, G.; Daniel, R.; Polle, A.; Reich, M. Fungal soil communities in a young transgenic poplar plantation form a rich reservoir for fungal root communities. Ecol. Evol. 2012, 2, 1935–1948. [Google Scholar] [CrossRef] [PubMed]
- Bahram, M.; Põlme, S.; Kõljalg, U.; Tedersoo, L. A single European aspen (Populus tremula) tree individual may potentially harbuor dozens of Cenococcum geophilum ITS genotypes and hundreds of species of ectomycorrhizal fungi. FEMS Microbiol. Ecol. 2011, 75, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Karlinski, L.; Rudawska, M.; Leski, T. The influence of host genotype and soil conditions on ectomycorrhizal community of poplar clones. Eur. J. Soil Biol. 2013, 58, 51–58. [Google Scholar] [CrossRef]
- Gamper, H.A.; Young, J.P.W.; Jones, D.L.; Hodge, A. Real-time PCR and microscopy: Are the two methods measuring the same unit of arbuscular mycorrhizal fungal abundance? Fungal Genet. Biol. 2008, 45, 581–596. [Google Scholar] [CrossRef] [PubMed]
- Agerer, R. Colour Atlas of Ectomycorrhizae; Einhorn-Verlag: Schwäbisch Gmünd, Germany, 1987–2006. [Google Scholar]
- Vierheilig, H.; Coughlan, A.P.; Wyss, U.; Piché, Y. Ink and vinegar a simple staining technique for arbuscular-mycorrhizal fungi. Appl. Environ. Microbiol. 1998, 64, 5004–5007. [Google Scholar] [PubMed]
- McGonigle, T.P.; Miller, M.H.; Evans, D.G.; Fairchild, G.L.; Swan, J.A. A new method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol. 1990, 115, 495–501. [Google Scholar] [CrossRef]
- Martin, K.J.; Rygiewicz, P.T. Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiol. 2005, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- NCBI. Available online: http://www.ncbi.nlm.nih.gov/ (accessed on 15 August 2014).
- UNITE. Available online: http://unite.ut.ee/ (accessed on 15 August 2014).
- Schwarzott, D.; Schüßler, A. A simple and reliable method for SSU rRNA gene DNA extraction, amplification, and cloning from single AM fungal spores. Mycorrhiza 2001, 10, 203–207. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, G.F.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Henry, M.; Stevens, H.; et al. Vegan: Community Ecology Package. R package version 2.0-10. Available online: http://CRAN.R-project.org/package=vegan (accessed on 15 August 2014).
- Bates, D.; Maechler, M.; Bolker, B.; Walker, S. lme4: Linear mixed-effect models using Eigen and S4. R package version 1.1-6. 2004. Available online: http://CRAN.R-project.org/package=lme (accessed on 1 October 2014).
- Hothorn, T.; Bretz, F.; Westfall, P. Simultaneous Inference in General Parametric. Models. Biometr. J. 2008, 50, 346–363. [Google Scholar] [CrossRef]
- Barker, S.J.; Duplessis, S.; Tagu, D. The application of genetic approaches for investigations of mycorrhizal symbioses. Plant Soil 2002, 244, 85–95. [Google Scholar] [CrossRef]
- Jumpponen, A.; Jones, K.L.; Mattox, J.D.; Yaege, C. Massively parallel 454-sequencing of fungal communities in Quercus spp. ectomycorrhizas indicates seasonal dynamics in urban and rural sites. Mol. Ecol. 2010, 19, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Baum, C.; Makeschin, F. Effects of nitrogen and phosphorus fertilization on mycorrhizal formation of two poplar clones (Populus trichocarpa and P. tremula × tremuloides). J. Plant Nutr. Soil Sci. 2000, 163, 491–497. [Google Scholar] [CrossRef]
- Treseder, K.K.; Allen, M.F. Direct N and P limitation of arbuscular mycorrhizal fungi: A model and field test. New Phytol. 2002, 155, 507–515. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gherghel, F.; Behringer, D.; Haubrich, S.; Schlauß, M.; Fey-Wagner, C.; Rexer, K.-H.; Janßen, A.; Kost, G. Former Land Use and Host Genotype Influence the Mycorrhizal Colonization of Poplar Roots. Forests 2014, 5, 2980-2995. https://doi.org/10.3390/f5122980
Gherghel F, Behringer D, Haubrich S, Schlauß M, Fey-Wagner C, Rexer K-H, Janßen A, Kost G. Former Land Use and Host Genotype Influence the Mycorrhizal Colonization of Poplar Roots. Forests. 2014; 5(12):2980-2995. https://doi.org/10.3390/f5122980
Chicago/Turabian StyleGherghel, Felicia, David Behringer, Stefanie Haubrich, Maren Schlauß, Christina Fey-Wagner, Karl-Heinz Rexer, Alwin Janßen, and Gerhard Kost. 2014. "Former Land Use and Host Genotype Influence the Mycorrhizal Colonization of Poplar Roots" Forests 5, no. 12: 2980-2995. https://doi.org/10.3390/f5122980
APA StyleGherghel, F., Behringer, D., Haubrich, S., Schlauß, M., Fey-Wagner, C., Rexer, K. -H., Janßen, A., & Kost, G. (2014). Former Land Use and Host Genotype Influence the Mycorrhizal Colonization of Poplar Roots. Forests, 5(12), 2980-2995. https://doi.org/10.3390/f5122980