Blood Mitochondrial DNA Content in HIV-Exposed Uninfected Children with Autism Spectrum Disorder
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Study Participants
2.3. Diagnosis and Severity of Autism Spectrum Disorder
2.4. Specimen Collection and Preparation
2.5. MtDNA Content Assay
2.6. Statistical Analyses
2.7. Sensitivity Analyses
3. Results
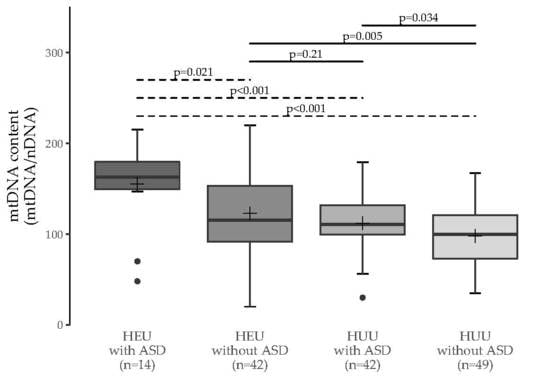
3.1. MtDNA Content Analyses
3.2. Multiple Group Comparisons
3.3. Sensitivity Analyses
3.3.1. Ethnicity
3.3.2. DNA Extract Dilution Factor
3.4. Clinical Outcomes
4. Discussion
Limitations
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Forbes, J.C.; Alimenti, A.M.; Singer, J.; Brophy, J.C.; Bitnun, A.; Samson, L.M.; Money, D.M.; Lee, T.C.K.; Lapointe, N.D.; Read, S.E. A national review of vertical HIV transmission. AIDS 2012, 26, 757–763. [Google Scholar] [CrossRef] [PubMed]
- Thorne, C.; Newell, M.-L. Safety of Agents Used to Prevent Mother-to-Child Transmission of HIV. Drug Saf. 2007, 30, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Thorne, C.; Newell, M.-L. The safety of antiretroviral drugs in pregnancy. Expert Opin. Drug Saf. 2005, 4, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Lewis, W.; Day, B.J.; Copeland, W.C. Mitochondrial toxicity of NRTI antiviral drugs: An integrated cellular perspective. Nat. Rev. Drug Discov. 2003, 2, 812–822. [Google Scholar] [CrossRef] [PubMed]
- Springer, P.E.; Slogrove, A.L.; Laughton, B.; Bettinger, J.A.; Saunders, H.H.; Molteno, C.D.; Kruger, M. Neurodevelopmental Outcome of HIV-exposed but uninfected infants in the Mother and Infants Health Study, Cape Town, South Africa. Trop. Med. Int. Health 2017, 23, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Côté, H.C.F.; Brumme, Z.L.; Craib, K.J.P.; Alexander, C.S.; Wynhoven, B.; Ting, L.; Wong, H.; Harris, M.; Harrigan, P.R.; O’Shaughnessy, M.V.; et al. Changes in mitochondrial DNA as a marker of nucleoside toxicity in HIV-infected patients. N. Engl. J. Med. 2002, 346, 811–820. [Google Scholar] [CrossRef]
- Hernàndez, S.; Morén, C.; López, M.; Coll, O.; Cardellach, F.; Gratacós, E.; Miró, O.; Garrabou, G. Perinatal outcomes, mitochondrial toxicity and apoptosis in HIV-treated pregnant women and in-utero-exposed newborn. AIDS 2012, 26, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Santini-Oliveira, M.; Grinsztejn, B. Adverse drug reactions associated with antiretroviral therapy during pregnancy. Expert Opin. Drug Saf. 2014, 13, 1623–1652. [Google Scholar] [CrossRef] [PubMed]
- Newell, M.-L.; Bunders, M.J. Safety of antiretroviral drugs in pregnancy and breastfeeding for mother and child. Curr. Opin. HIV AIDS 2013, 8, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Shiramizu, B.; Shikuma, K.M.; Kamemoto, L.; Gerschenson, M.; Erdem, G.; Pinti, M.; Cossarizza, A.; Shikuma, C. Placenta and cord blood mitochondrial DNA toxicity in HIV-infected women receiving nucleoside reverse transcriptase inhibitors during pregnancy. J. Acquir. Immune Defic. Syndr. 2003, 32, 370–374. [Google Scholar] [CrossRef] [PubMed]
- Decloedt, E.H.; Rosenkranz, B.; Maartens, G.; Joska, J. Central nervous system penetration of antiretroviral drugs: Pharmacokinetic, pharmacodynamic and pharmacogenomic considerations. Clin. Pharmacokinet. 2015, 54, 581–598. [Google Scholar] [CrossRef] [PubMed]
- Sibiude, J.; Warszawski, J.; Blanche, S. Tolerance of the newborn to antiretroviral drug exposure in utero. Expert Opin. Drug Saf. 2015, 14, 643–654. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.; Berk, M.; Walder, K.; Maes, M. Central pathways causing fatigue in neuro-inflammatory and autoimmune illnesses. BMC Med. 2015, 13, 28. [Google Scholar] [CrossRef]
- Giulivi, C.; Zhang, Y.-F.; Omanska-Klusek, A.; Ross-Inta, C.; Wong, S.; Hertz-Picciotto, I.; Tassone, F.; Pessah, I.N. Mitochondrial dysfunction in autism. JAMA 2010, 304, 2389–2396. [Google Scholar] [CrossRef] [PubMed]
- Frye, R.E.; Rossignol, D.A. Mitochondrial dysfunction can connect the diverse medical symptoms associated with autism spectrum disorders. Pediatr. Res. 2011, 69, 41R–47R. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, G.; Diogo, L.; Grazina, M.; Garcia, P.; Ataíde, A.; Marques, C.; Miguel, T.; Borges, L.; Vicente, A.; Oliveira, C. Mitochondrial dysfunction in autism spectrum disorders: A population-based study. Dev. Med. Child Neurol. 2005, 47, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Fatemi, S.H.; Sidwell, R.W.; Patterson, P.H. Maternal influenza infection causes marked behavioral and pharmacological changes in the offspring. J. Neurosci. 2003, 23, 297–302. [Google Scholar] [PubMed]
- Meyer, U.; Feldon, J.; Fatemi, S.H. In Vivo rodent models for the experimental investigation of prenatal immune activation effects in neurodevelopmental brain disorders. Neurosci. Biobehav. Rev. 2009, 33, 1061–1079. [Google Scholar] [CrossRef] [PubMed]
- Atladóttir, H.O.; Thorsen, P.; Østergaard, L.; Schendel, D.E.; Lemcke, S.; Abdallah, M.; Parner, E.T. Maternal infection requiring hospitalization during pregnancy and autism spectrum disorders. J. Autism Dev. Disord. 2010, 40, 1423–1430. [Google Scholar] [CrossRef] [PubMed]
- Matelski, L.; Van de Water, J. Risk factors in autism: Thinking outside the brain. J. Autoimmun. 2016, 67, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Côté, H.C.F.; Raboud, J.; Bitnun, A.; Alimenti, A.; Money, D.M.; Maan, E.; Costei, A.; Gadawski, I.; Diong, C.; Read, S.; et al. Perinatal exposure to antiretroviral therapy is associated with increased blood mitochondrial DNA levels and decreased mitochondrial gene expression in infants. J. Infect. Dis. 2008, 198, 851–859. [Google Scholar] [CrossRef]
- Barrientos, A.; Casademont, J.; Cardellach, F.; Estivill, X.; Urbano-Marquez, A.; Nunes, V. Reduced steady-state levels of mitochondrial RNA and increased mitochondrial DNA amount in human brain with aging. Brain Res. Mol. Brain Res. 1997, 52, 284–289. [Google Scholar] [CrossRef]
- Lagouge, M.; Larsson, N.-G. The role of mitochondrial DNA mutations and free radicals in disease and ageing. J. Intern. Med. 2013, 273, 529–543. [Google Scholar] [CrossRef] [PubMed]
- Autism and Developmental Disabilities Monitoring Network Surveillance Year 2010 Principal Investigators. Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years—Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2010. Morb. Mortal. Wkly. Rep. 2014, 63, 1–21. [Google Scholar]
- Bowes, J.; Brophy, J.C.; Samson, L.; Gentile, C. Pervasive Developmental Disorder in Antiretroviral- and HIV-Exposed, Uninfected Children [Abstract]. Can. J. Infect. Dis. Med. Microbiol. 2011, 22, 12B. [Google Scholar]
- Côté, H.C.F.; Soudeyns, H.; Thorne, A.; Alimenti, A.; Lamarre, V.; Maan, E.J.; Sattha, B.; Singer, J.; Lapointe, N.; Money, D.M.; et al. Leukocyte telomere length in HIV-infected and HIV-exposed uninfected children: Shorter telomeres with uncontrolled HIV viremia. PLoS ONE 2012, 7, e39266. [Google Scholar] [CrossRef]
- Zanet, D.L.; Saberi, S.; Oliveira, L.; Sattha, B.; Gadawski, I.; Côté, H.C.F. Blood and dried blood spot telomere length measurement by qPCR: Assay considerations. PLoS ONE 2013, 8, e57787. [Google Scholar] [CrossRef]
- Hsieh, A.Y.Y.; Saberi, S.; Ajaykumar, A.; Hukezalie, K.; Gadawski, I.; Sattha, B.; Côté, H.C.F. Optimization of a Relative Telomere Length Assay by Monochromatic Multiplex Real-Time Quantitative PCR on the LightCycler 480: Sources of Variability and Quality Control Considerations. J. Mol. Diagn. 2016, 18, 425–437. [Google Scholar] [CrossRef] [PubMed]
- Hurtado-Roca, Y.; Ledesma, M.; Gonzalez-Lazaro, M.; Moreno-Loshuertos, R.; Fernandez-Silva, P.; Enriquez, J.A.; Laclaustra, M. Adjusting MtDNA Quantification in Whole Blood for Peripheral Blood Platelet and Leukocyte Counts. PLoS ONE 2016, 11, e0163770. [Google Scholar] [CrossRef] [PubMed]
- Urata, M.; Koga-Wada, Y.; Kayamori, Y.; Kang, D. Platelet contamination causes large variation as well as overestimation of mitochondrial DNA content of peripheral blood mononuclear cells. Ann. Clin. Biochem. 2008, 45, 513–514. [Google Scholar] [CrossRef] [PubMed]
- Banas, B.; Kost, B.P.; Goebel, F.D. Platelets, a typical source of error in real-time PCR quantification of mitochondrial DNA content in human peripheral blood cells. Eur. J. Med. Res. 2004, 9, 371–377. [Google Scholar] [PubMed]
- McComsey, G.A.; Kang, M.; Ross, A.C.; Lebrecht, D.; Livingston, E.; Melvin, A.; Hitti, J.; Cohn, S.E.; Walker, U.A. Increased mtDNA Levels Without Change in Mitochondrial Enzymes in Peripheral Blood Mononuclear Cells of Infants Born to HIV-Infected Mothers on Antiretroviral Therapy. HIV Clin. Trials 2015, 9, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Ross, A.C.; Leong, T.; Avery, A.; Castillo-Duran, M.; Bonilla, H.; Lebrecht, D.; Walker, U.A.; Storer, N.; Labbato, D.; Khaitan, A.; et al. Effects of in utero antiretroviral exposure on mitochondrial DNA levels, mitochondrial function and oxidative stress. HIV Med. 2012, 13, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Aldrovandi, G.M.; Chu, C.; Shearer, W.T.; Li, D.; Walter, J.; Thompson, B.; McIntosh, K.; Foca, M.; Meyer, W.A.; Ha, B.F.; et al. Antiretroviral exposure and lymphocyte mtDNA content among uninfected infants of HIV-1-infected women. Pediatrics 2009, 124, e1189–e1197. [Google Scholar] [CrossRef] [PubMed]
- Poirier, M.C.; Divi, R.L.; Al-Harthi, L.; Olivero, O.A.; Nguyen, V.; Walker, B.; Landay, A.L.; Walker, V.E.; Charurat, M.; Blattner, W.A. Long-Term Mitochondrial Toxicity in HIV-Uninfected Infants Born to HIV-Infected Mothers. JAIDS J. Acquir. Immune Defic. Syndr. 2003, 33, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Montaner, J.S.G.; Côté, H.C.F.; Harris, M.; Hogg, R.S.; Yip, B.; Chan, J.W.; Harrigan, P.R.; O’Shaughnessy, M.V. Mitochondrial toxicity in the era of HAART: Evaluating venous lactate and peripheral blood mitochondrial DNA in HIV-infected patients taking antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 2003, 34 (Suppl. 1), S85–S90. [Google Scholar] [CrossRef] [PubMed]
- Chalkia, D.; Singh, L.N.; Leipzig, J.; Lvova, M.; Derbeneva, O.; Lakatos, A.; Hadley, D.; Hakonarson, H.; Wallace, D.C. Association Between Mitochondrial DNA Haplogroup Variation and Autism Spectrum Disorders. JAMA Psychiatry 2017, 74, 1161. [Google Scholar] [CrossRef] [PubMed]
- Rao, R.; Rivers, A.; Rahimi, A.; Wooldridge, R.; Rao, M.; Leitch, M.; Euhus, D.; Haley, B.B. Genetic Ancestry using Mitochondrial DNA in patients with Triple-negative breast cancer (GAMiT study). Cancer 2017, 123, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Cardena, M.M.; Ribeiro-Dos-Santos, A.; Santos, S.; Mansur, A.J.; Pereira, A.C.; Fridman, C. Assessment of the relationship between self-declared ethnicity, mitochondrial haplogroups and genomic ancestry in Brazilian individuals. PLoS ONE 2013, 8, e62005. [Google Scholar] [CrossRef] [PubMed]
- Salas, A.; Fachal, L.; Marcos-Alonso, S.; Vega, A.; Martinón-Torres, F.; Grupo de investigación ESIGEM (Estudio Sobre la Influencia Genética en la Enfermedad Meningocócica). Investigating the role of mitochondrial haplogroups in genetic predisposition to meningococcal disease. PLoS ONE 2009, 4, e8347. [Google Scholar] [CrossRef] [PubMed]
- Salas, A.; Acosta, A.; Álvarez-Iglesias, V.; Cerezo, M.; Phillips, C.; Lareu, M.V.; Carracedo, Á. The mtDNA ancestry of admixed Colombian populations. Am. J. Hum. Biol. 2008, 20, 584–591. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Li, Z.; He, Y.; Zhang, F.; Li, H.; Liao, Y.; Wei, Z.; Wan, G.; Xiang, X.; Hu, M.; et al. Elevated mitochondrial DNA copy number in peripheral blood cells is associated with childhood autism. BMC Psychiatry 2015, 15, 50. [Google Scholar] [CrossRef] [PubMed]
- James, S.J.; Cutler, P.; Melnyk, S.; Jernigan, S.; Janak, L.; Gaylor, D.W.; Neubrander, J.A. Metabolic biomarkers of increased oxidative stress and impaired methylation capacity in children with autism. Am. J. Clin. Nutr. 2004, 80, 1611–1617. [Google Scholar] [CrossRef] [PubMed]
- Rossignol, D.A.; Frye, R.E. Mitochondrial dysfunction in autism spectrum disorders: A systematic review and meta-analysis. Mol. Psychiatry 2012, 17, 290–314. [Google Scholar] [CrossRef] [PubMed]
- Brogly, S.B.; Ylitalo, N.; Mofenson, L.M.; Oleske, J.; van Dyke, R.; Crain, M.J.; Abzug, M.J.; Brady, M.; Jean-Philippe, P.; Hughes, M.D.; et al. In utero nucleoside reverse transcriptase inhibitor exposure and signs of possible mitochondrial dysfunction in HIV-uninfected children. AIDS 2007, 21, 929–938. [Google Scholar] [CrossRef] [PubMed]
- Mofenson, L.M. Editorial commentary: New challenges in the elimination of pediatric HIV infection: The expanding population of HIV-exposed but uninfected children. Clin. Infect. Dis. 2015, 60, 1357–1360. [Google Scholar] [CrossRef] [PubMed]
- Harris, P.A.; Taylor, R.; Thielke, R.; Payne, J.; Gonzalez, N.; Conde, J.G. Research electronic data capture (REDCap)—A metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 2009, 42, 377–381. [Google Scholar] [CrossRef] [PubMed]




| Characteristic | HEU with ASD n = 14 | HEU without ASD n = 42 | HUU with ASD n = 42 | HUU without ASD n = 51 |
|---|---|---|---|---|
| Male sex | 10/14 (71) | 30/42 (71) | 30/42 (71) | 36/51 (71) |
| Age at blood collection (years) | 6 (4–8) [2–16] | 6 (4–8) [2–16] | 6 (4–8) [2–16] | 6 (4–9) [2–16] |
| Ethnicity | ||||
| Black/African-Canadian | 11/14 (79) | 33/42 (79) | 6/42 (14) | 0/51 |
| White/Caucasian | 3/14 (21) | 9/42 (21) | 22/42 (52) | 5/51 (10) |
| Asian/South Asian | 0/14 (0) | 0/42 (0) | 10/42 (24) | 4/51 (8) |
| Indigenous | 0/14 (0) | 0/42 (0) | 4/42 (10) | 0/51 (0) |
| Unknown (anonymous) | 0/14 (0) | 0/42 (0) | 0/42 (0) | 42/51 (82) |
| Maternal age at birth (years) | 30 (24–32) [18–42] | 33 (31–36) [25–42] | 31 (28–35) [18–42] | 27 (26–36) [21–39] |
| Paternal age at birth (years) | 35 (29–42) [25–44] | 37 (34–41) [24–63] | 35 (30–38) [20–51] | 30 (28–38) [26–44] |
| Characteristic | HEU with ASD n = 14 | HEU without ASD n = 42 | HUU with ASD n = 42 | HUU without ASD n = 51 |
|---|---|---|---|---|
| Developmental disorders/delays | ||||
| Intellectual disabilities | 9/14 (64) | 0/42 (0) | 22/42 (52) | 0/9 (0) |
| Language delay | 2/14 (14) | 2/42 (5) | 6/42 (14) | 0/9 (0) |
| Unknown or unable to assess | 2/14 (14) | 0/42 (0) | 4/42 (10) | 42/51 (82) |
| Severity of ASD symptoms | ||||
| Mild or mild/moderate | 5/14 (36) | - | 7/42 (17) | - |
| Moderate | 1/14 (7) | - | 4/42 (10) | - |
| Moderate/severe or severe | 7/14 (50) | - | 5/42 (12) | - |
| Unable to assess | 0/14 (0) | - | 13/42 (31) | - |
| Not specified | 1/14 (7) | - | 13/42 (31) | - |
| History of seizures/epilepsy (ever) | 2/14 (14) | 0/42 (0) | 5/42 (12) | 0/9 (0) |
| Low muscle tone | 3/14 (21) | 2/42 (5) | 14/42 (33) | 1/9 (11) |
| Chronic gastrointestinal disorders | 10/14 (71) | 9/42 (21) | 7/42 (17) | 0/9 (0) |
| Maternal ARV regimen during pregnancy 1 | ||||
| Dual NRTIs+PI | ||||
| ZDV + 3TC + PI | 5/14 (36) | 19/42 (45) | - | - |
| ABC + 3TC + PI | 0/14 | 8/42 (19) | - | - |
| TDF + FTC + PI | 2/14 (14) | 3/42 (7) | - | - |
| Other combinations + PI | 1/14 (7) | 4/42 (10) | ||
| Dual NRTIs + NNRTI | 1/14 (7) | 2/42 (5) | - | - |
| NRTI + NNRTI + PI | 0/14 (0) | 2/42 (5) | ||
| Other | 1/14 (7) | 1/42 (2) | - | - |
| None | 3/14 (21) | 2/42 (5) | - | - |
| Unknown | 1/14 (7) | 1/42 (2) | - | - |
| Infant received ZDV prophylaxis | 13/14 (93) | 42/42 (100) | - | - |
| Length of in utero ARV exposure (weeks) 2 | 11 (6–35) [0–38] | 33 (20–39) [0–41] | - | - |
| Length of neonatal ZDV prophylaxis (weeks) | 6 (4–6) [0–7] | 6 (6–6) [4–7] | - | - |
| HEU with ASD | HEU without ASD | HUU with ASD | |
|---|---|---|---|
| HEU without ASD | 0.011 (0.021) | - | - |
| HUU with ASD | 0.002 (<0.001) | 0.38 (0.21) | - |
| HUU without ASD | <0.001 (<0.001) | 0.006 (0.005) | 0.063 (0.034) |
| Asian/South Asian n = 10 | White/Caucasian n = 20 | Black/African-Canadian n = 20 | |
|---|---|---|---|
| Male sex | 5 (50) | 10 (50) | 10 (50) |
| Age at blood collection (years) | 1.3 (0.6–2.5) [0.3–15.1] | 1.4 (0.5–2.6) [0.2–15.7] | 1.3 (0.5–2.6) [0.3–15.6] |
| MtDNA content | 128 (115–196) [90–203] | 160 (99–191) [54–259] | 169 (133–192) [62–268] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Budd, M.A.; Calli, K.; Samson, L.; Bowes, J.; Hsieh, A.Y.Y.; Forbes, J.C.; Bitnun, A.; Singer, J.; Kakkar, F.; Alimenti, A.; et al. Blood Mitochondrial DNA Content in HIV-Exposed Uninfected Children with Autism Spectrum Disorder. Viruses 2018, 10, 77. https://doi.org/10.3390/v10020077
Budd MA, Calli K, Samson L, Bowes J, Hsieh AYY, Forbes JC, Bitnun A, Singer J, Kakkar F, Alimenti A, et al. Blood Mitochondrial DNA Content in HIV-Exposed Uninfected Children with Autism Spectrum Disorder. Viruses. 2018; 10(2):77. https://doi.org/10.3390/v10020077
Chicago/Turabian StyleBudd, Matthew A., Kristina Calli, Lindy Samson, Jennifer Bowes, Anthony Y. Y. Hsieh, John C. Forbes, Ari Bitnun, Joel Singer, Fatima Kakkar, Ariane Alimenti, and et al. 2018. "Blood Mitochondrial DNA Content in HIV-Exposed Uninfected Children with Autism Spectrum Disorder" Viruses 10, no. 2: 77. https://doi.org/10.3390/v10020077
APA StyleBudd, M. A., Calli, K., Samson, L., Bowes, J., Hsieh, A. Y. Y., Forbes, J. C., Bitnun, A., Singer, J., Kakkar, F., Alimenti, A., Maan, E. J., Lewis, M. E. S., Gentile, C., Côté, H. C. F., & Brophy, J. C. (2018). Blood Mitochondrial DNA Content in HIV-Exposed Uninfected Children with Autism Spectrum Disorder. Viruses, 10(2), 77. https://doi.org/10.3390/v10020077