The Enigmatic Alphavirus Non-Structural Protein 3 (nsP3) Revealing Its Secrets at Last
Abstract
:1. Introduction
2. Expression of the Non-Structural Polyprotein Is Regulated by the Presence of Opal Stop Codon at the nsP3/4 Junction in Some Alphaviruses
3. A Degradation Signal at the Extreme C-Terminus of nsP3 Regulates Its Expression
4. Localization of nsP3 in Infected Cells
5. NsP3 as a Vector Specificity Determinant
6. NsP3 Is a Major Determinant of Neurovirulence for Some Alphaviruses
7. NsP3 Is a Hub for Multiple Host Protein Interactions
8. The Macro Domain
9. The Central Alphavirus-Unique Domain (AUD) Is a Zinc-Binding Domain (ZBD)
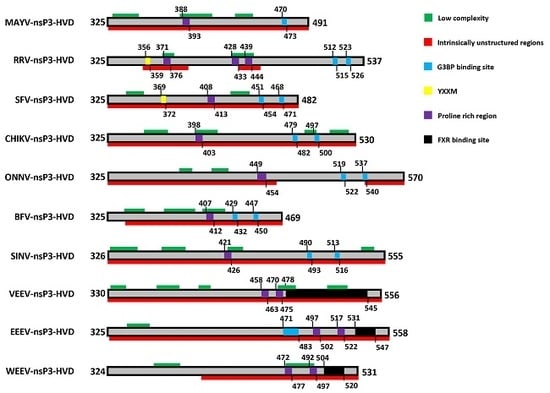
10. The Hypervariable Domain (HVD)
11. The HVD Is Phosphorylated
12. A YXXM Motif Directs Hyperactivation of the PI3K-Akt-mTOR Pathway
13. A Proline-Rich Region within the HVD Mediates Binding to Amphiphysin Proteins
14. The HVD Contains Repeat Motifs Which Direct Binding to Host Stress Granule Proteins
15. Conclusions
Acknowledgments
Conflicts of Interest
References
- Ng, L.F.P. Immunopathology of Chikungunya Virus Infection: Lessons Learned from Patients and Animal Models. Annu. Rev. Virol. 2017, 4, 413–427. [Google Scholar] [CrossRef] [PubMed]
- Josseran, L.; Paquet, C.; Zehgnoun, A.; Caillere, N.; Le Tertre, A.; Solet, J.L.; Ledrans, M. Chikungunya disease outbreak, Reunion Island. Emerg. Infect. Dis. 2006, 12, 1994–1995. [Google Scholar] [CrossRef] [PubMed]
- Mavalankar, D.; Shastri, P.; Bandyopadhyay, T.; Parmar, J.; Ramani, K.V. Increased mortality rate associated with chikungunya epidemic, Ahmedabad, India. Emerg. Infect. Dis. 2008, 14, 412–415. [Google Scholar] [CrossRef] [PubMed]
- Morrison, T.E. Reemergence of chikungunya virus. J. Virol. 2014, 88, 11644–11647. [Google Scholar] [CrossRef] [PubMed]
- Weaver, S.C.; Forrester, N.L. Chikungunya: Evolutionary history and recent epidemic spread. Antivir. Res. 2015, 120, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, M.L.; Figueiredo, L.T. Emerging alphaviruses in the Americas: Chikungunya and Mayaro. Rev. Soc. Bras. Med. Trop. 2014, 47, 677–683. [Google Scholar] [CrossRef] [PubMed]
- Lwande, O.W.; Obanda, V.; Bucht, G.; Mosomtai, G.; Otieno, V.; Ahlm, C.; Evander, M. Global emergence of Alphaviruses that cause arthritis in humans. Infect. Ecol. Epidemiol. 2015, 5, 29853. [Google Scholar] [CrossRef] [PubMed]
- DeTulleo, L.; Kirchhausen, T. The clathrin endocytic pathway in viral infection. EMBO J. 1998, 17, 4585–4593. [Google Scholar] [CrossRef] [PubMed]
- Dubuisson, J.; Rice, C.M. Sindbis virus attachment: Isolation and characterization of mutants with impaired binding to vertebrate cells. J. Virol. 1993, 67, 3363–3374. [Google Scholar] [PubMed]
- Vasiljeva, L.; Valmu, L.; Kaariainen, L.; Merits, A. Site-specific protease activity of the carboxyl-terminal domain of Semliki Forest virus replicase protein nsP2. J. Biol. Chem. 2001, 276, 30786–30793. [Google Scholar] [CrossRef] [PubMed]
- Kaariainen, L.; Ahola, T. Functions of alphavirus nonstructural proteins in RNA replication. Prog. Nucleic Acid. Res. Mol. Biol. 2002, 71, 187–222. [Google Scholar] [PubMed]
- Strauss, J.H.; Strauss, E.G. The alphaviruses: Gene expression, replication and evolution. Microbiol. Rev. 1994, 58, 491–562. [Google Scholar] [PubMed]
- Spuul, P.; Balistreri, G.; Kaariainen, L.; Ahola, T. Phosphatidylinositol 3-kinase-, actin- and microtubule-dependent transport of Semliki Forest Virus replication complexes from the plasma membrane to modified lysosomes. J. Virol. 2010, 84, 7543–7557. [Google Scholar] [CrossRef] [PubMed]
- Frolova, E.I.; Gorchakov, R.; Pereboeva, L.; Atasheva, S.; Frolov, I. Functional Sindbis virus replicative complexes are formed at the plasma membrane. J. Virol. 2010, 84, 11679–11695. [Google Scholar] [CrossRef] [PubMed]
- Froshauer, S.; Kartenbeck, J.; Helenius, A. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 1988, 107 Pt 1, 2075–2086. [Google Scholar] [CrossRef] [PubMed]
- Shirako, Y.; Strauss, J.H. Regulation of Sindbis virus RNA replication: Uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J. Virol. 1994, 68, 1874–1885. [Google Scholar] [PubMed]
- Peranen, J.; Rikkonen, M.; Liljestrom, P.; Kaariainen, L. Nuclear localization of Semliki Forest virus-specific nonstructural protein nsP2. J. Virol. 1990, 64, 1888–1896. [Google Scholar] [PubMed]
- Gorchakov, R.; Garmashova, N.; Frolova, E.; Frolov, I. Different types of nsP3-containing protein complexes in Sindbis virus-infected cells. J. Virol. 2008, 82, 10088–10101. [Google Scholar] [CrossRef] [PubMed]
- Peranen, J.; Laakkonen, P.; Hyvonen, M.; Kaariainen, L. The alphavirus replicase protein nsP1 is membrane-associated and has affinity to endocytic organelles. Virology 1995, 208, 610–620. [Google Scholar] [CrossRef] [PubMed]
- Ahola, T.; Kaariainen, L. Reaction in alphavirus mRNA capping: Formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc. Natl. Acad. Sci. USA 1995, 92, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Salonen, A.; Vasiljeva, L.; Merits, A.; Magden, J.; Jokitalo, E.; Kaariainen, L. Properly folded nonstructural polyprotein directs the semliki forest virus replication complex to the endosomal compartment. J. Virol. 2003, 77, 1691–1702. [Google Scholar] [CrossRef] [PubMed]
- Gomez de Cedron, M.; Ehsani, N.; Mikkola, M.L.; Garcia, J.A.; Kaariainen, L. RNA helicase activity of Semliki Forest virus replicase protein NSP2. FEBS Lett. 1999, 448, 19–22. [Google Scholar] [CrossRef]
- Tomar, S.; Hardy, R.W.; Smith, J..; Kuhn, R.J. Catalytic core of alphavirus nonstructural protein nsP4 possesses terminal adenylyltransferase activity. J. Virol. 2006, 80, 9962–9969. [Google Scholar] [CrossRef] [PubMed]
- Hahn, Y.S.; Strauss, E.G.; Strauss, J.H. Mapping of RNA- temperature-sensitive mutants of Sindbis virus: Assignment of complementation groups A, B and G to nonstructural proteins. J. Virol. 1989, 63, 3142–3150. [Google Scholar] [PubMed]
- LaStarza, M.W.; Lemm, J.A.; Rice, C.M. Genetic analysis of the nsP3 region of Sindbis virus: Evidence for roles in minus-strand and subgenomic RNA synthesis. J. Virol. 1994, 68, 5781–5791. [Google Scholar] [PubMed]
- Wang, Y.F.; Sawicki, S.G.; Sawicki, D.L. Alphavirus nsP3 functions to form replication complexes transcribing negative-strand RNA. J. Virol. 1994, 68, 6466–6475. [Google Scholar] [PubMed]
- Keranen, S.; Ruohonen, L. Nonstructural proteins of Semliki Forest virus: Synthesis, processing and stability in infected cells. J. Virol. 1983, 47, 505–515. [Google Scholar] [PubMed]
- Lachmi, B.E.; Kaariainen, L. Sequential translation of nonstructural proteins in cells infected with a Semliki Forest virus mutant. Proc. Natl. Acad. Sci. USA 1976, 73, 1936–1940. [Google Scholar] [CrossRef] [PubMed]
- Takkinen, K. Complete nucleotide sequence of the nonstructural protein genes of Semliki Forest virus. Nucleic Acids Res. 1986, 14, 5667–5682. [Google Scholar] [CrossRef] [PubMed]
- Lopez, S.; Bell, J.R.; Strauss, E.G.; Strauss, J.H. The nonstructural proteins of Sindbis virus as studied with an antibody specific for the C terminus of the nonstructural readthrough polyprotein. Virology 1985, 141, 235–247. [Google Scholar] [CrossRef]
- Strauss, E.G.; Levinson, R.; Rice, C.M.; Dalrymple, J.; Strauss, J.H. Nonstructural proteins nsP3 and nsP4 of Ross River and O’Nyong-nyong viruses: Sequence and comparison with those of other alphaviruses. Virology 1988, 164, 265–274. [Google Scholar] [CrossRef]
- Lanciotti, R.S.; Ludwig, M.L.; Rwaguma, E.B.; Lutwama, J.J.; Kram, T.M.; Karabatsos, N.; Cropp, B.C.; Miller, B.R. Emergence of epidemic O’Nyong-nyong fever in Uganda after a 35-year absence: Genetic characterization of the virus. Virology 1998, 252, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Myles, K.M.; Kelly, C.L.; Ledermann, J.P.; Powers, A.M. Effects of an opal termination codon preceding the nsP4 gene sequence in the O'Nyong-Nyong virus genome on Anopheles gambiae infectivity. J. Virol. 2006, 80, 4992–4997. [Google Scholar] [CrossRef] [PubMed]
- Stapleford, K.A.; Moratorio, G.; Henningsson, R.; Chen, R.; Matheus, S.; Enfissi, A.; Weissglas-Volkov, D.; Isakov, O.; Blanc, H.; Mounce, B.C.; et al. Whole-Genome Sequencing Analysis from the Chikungunya Virus Caribbean Outbreak Reveals Novel Evolutionary Genomic Elements. PLoS Negl. Trop. Dis. 2016, 10, e0004402. [Google Scholar] [CrossRef] [PubMed]
- Teo, T.H.; Her, Z.; Tan, J.J.; Lum, F.M.; Lee, W.W.; Chan, Y.H.; Ong, R.Y.; Kam, Y.W.; Leparc-Goffart, I.; Gallian, P.; et al. Caribbean and La Reunion Chikungunya Virus Isolates Differ in Their Capacity To Induce Proinflammatory Th1 and NK Cell Responses and Acute Joint Pathology. J. Virol. 2015, 89, 7955–7969. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.C.; Kam, Y.W.; Lin, R.T.; Ng, M.M.; Ng, L.F.; Chu, J.J. Comparative analysis of the genome sequences and replication profiles of chikungunya virus isolates within the East, Central and South African (ECSA) lineage. Virol. J. 2013, 10, 169. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.E.; Long, K.M.; Whitmore, A.C.; Sanders, W.; Thurlow, L.R.; Brown, J.A.; Morrison, C.R.; Vincent, H.; Peck, K.M.; Browning, C.; et al. Disruption of the Opal Stop Codon Attenuates Chikungunya Virus-Induced Arthritis and Pathology. mBio 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Tuittila, M.T.; Santagati, M.G.; Roytta, M.; Maatta, J.A.; Hinkkanen, A.E. Replicase complex genes of Semliki Forest virus confer lethal neurovirulence. J. Virol. 2000, 74, 4579–4589. [Google Scholar] [CrossRef] [PubMed]
- Tuittila, M.; Hinkkanen, A.E. Amino acid mutations in the replicase protein nsP3 of Semliki Forest virus cumulatively affect neurovirulence. J. Gen. Virol. 2003, 84 Pt 6, 1525–1533. [Google Scholar] [CrossRef] [PubMed]
- Varjak, M.; Zusinaite, E.; Merits, A. Novel functions of the alphavirus nonstructural protein nsP3 C-terminal region. J. Virol. 2010, 84, 2352–2364. [Google Scholar] [CrossRef] [PubMed]
- Saul, S.; Ferguson, M.; Cordonin, C.; Fragkoudis, R.; Ool, M.; Tamberg, N.; Sherwood, K.; Fazakerley, J.K.; Merits, A. Differences in Processing Determinants of Nonstructural Polyprotein and in the Sequence of Nonstructural Protein 3 Affect Neurovirulence of Semliki Forest Virus. J. Virol. 2015, 89, 11030–11045. [Google Scholar] [CrossRef] [PubMed]
- Albulescu, I.C.; Tas, A.; Scholte, F.E.; Snijder, E.J.; van Hemert, M.J. An in vitro assay to study chikungunya virus RNA synthesis and the mode of action of inhibitors. J. Gen. Virol. 2014, 95 Pt 12, 2683–2692. [Google Scholar] [CrossRef] [PubMed]
- Peranen, J.; Takkinen, K.; Kalkkinen, N.; Kaariainen, L. Semliki Forest virus-specific non-structural protein nsP3 is a phosphoprotein. J. Gen. Virol. 1988, 69 Pt 9, 2165–2178. [Google Scholar] [CrossRef] [PubMed]
- Ranki, M.; Kaariainen, L. Solubilized RNA replication complex from Semliki Forest virus-infected cells. Virology 1979, 98, 298–307. [Google Scholar] [CrossRef]
- Pietila, M.K.; van Hemert, M.J.; Ahola, T. Purification of highly active alphavirus replication complexes demonstrates altered fractionation of multiple cellular membranes. J. Virol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Cristea, I.M.; Carroll, J.W.; Rout, M.P.; Rice, C.M.; Chait, B.T.; MacDonald, M.R. Tracking and elucidating alphavirus-host protein interactions. J. Biol. Chem. 2006, 281, 30269–30278. [Google Scholar] [CrossRef] [PubMed]
- Kujala, P.; Ikaheimonen, A.; Ehsani, N.; Vihinen, H.; Auvinen, P.; Kaariainen, L. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 2001, 75, 3873–3884. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Li, G. Recombinant Sindbis virus expressing functional GFP in the nonstructural protein nsP3. Gene Ther. Mol. Biol. 2005, 9, 317–324. [Google Scholar]
- Vihinen, H.; Ahola, T.; Tuittila, M.; Merits, A.; Kaariainen, L. Elimination of phosphorylation sites of Semliki Forest virus replicase protein nsP3. J. Biol. Chem. 2001, 276, 5745–5752. [Google Scholar] [CrossRef] [PubMed]
- Peränen, J.; Kaariainen, L. Biogenesis of type I cytopathic vacuoles in Semliki Forest virus-infected BHK cells. J. Virol. 1991, 65, 1623–1627. [Google Scholar] [PubMed]
- Foy, N.J.; Akhrymuk, M.; Akhrymuk, I.; Atasheva, S.; Bopda-Waffo, A.; Frolov, I.; Frolova, E.I. Hypervariable domains of nsP3 proteins of New World and Old World alphaviruses mediate formation of distinct, virus-specific protein complexes. J. Virol. 2013, 87, 1997–2010. [Google Scholar] [CrossRef] [PubMed]
- Panas, M.D.; Ahola, T.; McInerney, G.M. The C-terminal repeat domains of nsP3 from the Old World alphaviruses bind directly to G3BP. J. Virol. 2014, 88, 5888–5893. [Google Scholar] [CrossRef] [PubMed]
- Panas, M.D.; Varjak, M.; Lulla, A.; Eng, K.E.; Merits, A.; Karlsson Hedestam, G.B.; McInerney, G.M. Sequestration of G3BP coupled with efficient translation inhibits stress granules in Semliki Forest virus infection. Mol. Biol. Cell 2012, 23, 4701–4712. [Google Scholar] [CrossRef] [PubMed]
- Sreejith, R.; Rana, J.; Dudha, N.; Kumar, K.; Gabrani, R.; Sharma, S.K.; Gupta, A.; Vrati, S.; Chaudhary, V.K.; Gupta, S. Mapping interactions of Chikungunya virus nonstructural proteins. Virus Res. 2012, 169, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Saxton-Shaw, K.D.; Ledermann, J.P.; Borland, E.M.; Stovall, J.L.; Mossel, E.C.; Singh, A.J.; Wilusz, J.; Powers, A.M. O’nyong nyong virus molecular determinants of unique vector specificity reside in non-structural protein 3. PLoS Negl. Trop. Dis. 2013, 7, e1931. [Google Scholar] [CrossRef] [PubMed]
- Lastarza, M.W.; Grakoui, A.; Rice, C.M. Deletion and duplication mutations in the C-terminal nonconserved region of Sindbis virus nsP3: Effects on phosphorylation and on virus replication in vertebrate and invertebrate cells. Virology 1994, 202, 224–232. [Google Scholar] [CrossRef] [PubMed]
- Mathur, K.; Anand, A.; Dubey, S.K.; Sanan-Mishra, N.; Bhatnagar, R.K.; Sunil, S. Analysis of chikungunya virus proteins reveals that non-structural proteins nsP2 and nsP3 exhibit RNA interference (RNAi) suppressor activity. Sci. Rep. 2016, 6, 38065. [Google Scholar] [CrossRef] [PubMed]
- Suthar, M.S.; Shabman, R.; Madric, K.; Lambeth, C.; Heise, M.T. Identification of adult mouse neurovirulence determinants of the Sindbis virus strain AR86. J. Virol. 2005, 79, 4219–4228. [Google Scholar] [CrossRef] [PubMed]
- Atkins, G.J.; Sheahan, B.J. Molecular determinants of alphavirus neuropathogenesis in mice. J. Gen. Virol. 2016, 97, 1283–1296. [Google Scholar] [CrossRef] [PubMed]
- Frolova, E.; Gorchakov, R.; Garmashova, N.; Atasheva, S.; Vergara, L.A.; Frolov, I. Formation of nsP3-specific protein complexes during Sindbis virus replication. J. Virol. 2006, 80, 4122–4134. [Google Scholar] [CrossRef] [PubMed]
- Varjak, M.; Saul, S.; Arike, L.; Lulla, A.; Peil, L.; Merits, A. Magnetic fractionation and proteomic dissection of cellular organelles occupied by the late replication complexes of Semliki Forest virus. J. Virol. 2013, 87, 10295–10312. [Google Scholar] [CrossRef] [PubMed]
- Neuvonen, M.; Kazlauskas, A.; Martikainen, M.; Hinkkanen, A.; Ahola, T.; Saksela, K. SH3 domain-mediated recruitment of host cell amphiphysins by alphavirus nsP3 promotes viral RNA replication. PLoS Pathog. 2011, 7, e1002383. [Google Scholar] [CrossRef] [PubMed]
- Mazzon, M.; Castro, C.; Thaa, B.; Liu, L.; Mutso, M.; Liu, X.; Mahalingam, S.; Griffin, J.L.; Marsh, M.; McInerney, G.M. Alphavirus-induced hyperactivation of PI3K/AKT directs pro-viral metabolic changes. PLoS Pathog. 2018, 14, e1006835. [Google Scholar] [CrossRef] [PubMed]
- Park, E.; Griffin, D.E. Interaction of Sindbis virus non-structural protein 3 with poly(ADP-ribose) polymerase 1 in neuronal cells. J. Gen. Virol. 2009, 90 Pt 9, 2073–2080. [Google Scholar] [CrossRef] [PubMed]
- Amaya, M.; Voss, K.; Sampey, G.; Senina, S.; de la Fuente, C.; Mueller, C.; Calvert, V.; Kehn-Hall, K.; Carpenter, C.; Kashanchi, F.; et al. The role of IKKbeta in Venezuelan equine encephalitis virus infection. PLoS ONE 2014, 9, e86745. [Google Scholar] [CrossRef] [PubMed]
- Frolov, I.; Kim, D.Y.; Akhrymuk, M.; Mobley, J.A.; Frolova, E.I. Hypervariable Domain of Eastern Equine Encephalitis Virus nsP3 Redundantly Utilizes Multiple Cellular Proteins for Replication Complex Assembly. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.Y.; Reynaud, J.M.; Rasalouskaya, A.; Akhrymuk, I.; Mobley, J.A.; Frolov, I.; Frolova, E.I. New World and Old World Alphaviruses Have Evolved to Exploit Different Components of Stress Granules, FXR and G3BP Proteins, for Assembly of Viral Replication Complexes. PLoS Pathog. 2016, 12, e1005810. [Google Scholar] [CrossRef] [PubMed]
- Panas, M.D.; Schulte, T.; Thaa, B.; Sandalova, T.; Kedersha, N.; Achour, A.; McInerney, G.M. Viral and Cellular Proteins Containing FGDF Motifs Bind G3BP to Block Stress Granule Formation. PLoS Pathog. 2015, 11, e1004659. [Google Scholar] [CrossRef] [PubMed]
- Tossavainen, H.; Aitio, O.; Hellman, M.; Saksela, K.; Permi, P. Structural Basis of the High Affinity Interaction between the Alphavirus Nonstructural Protein-3 (nsP3) and the SH3 Domain of Amphiphysin-2. J. Biol. Chem. 2016, 291, 16307–16317. [Google Scholar] [CrossRef] [PubMed]
- Amaya, M.; Brooks-Faulconer, T.; Lark, T.; Keck, F.; Bailey, C.; Raman, V.; Narayanan, A. Venezuelan equine encephalitis virus non-structural protein 3 (nsP3) interacts with RNA helicases DDX1 and DDX3 in infected cells. Antivir. Res. 2016, 131, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Fros, J.J.; Geertsema, C.; Zouache, K.; Baggen, J.; Domeradzka, N.; van Leeuwen, D.M.; Flipse, J.; Vlak, J.M.; Failloux, A.B.; Pijlman, G.P. Mosquito Rasputin interacts with chikungunya virus nsP3 and determines the infection rate in Aedes albopictus. Parasites Vectors 2015, 8, 464. [Google Scholar] [CrossRef] [PubMed]
- Burnham, A.J.; Gong, L.; Hardy, R.W. Heterogeneous nuclear ribonuclear protein K interacts with Sindbis virus nonstructural proteins and viral subgenomic mRNA. Virology 2007, 367, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Rathore, A.P.; Haystead, T.; Das, P.K.; Merits, A.; Ng, M.L.; Vasudevan, S.G. Chikungunya virus nsP3 & nsP4 interacts with HSP-90 to promote virus replication: HSP-90 inhibitors reduce CHIKV infection and inflammation in vivo. Antivir. Res. 2014, 103, 7–16. [Google Scholar] [PubMed]
- Reid, S.P.; Tritsch, S.R.; Kota, K.; Chiang, C.Y.; Dong, L.; Kenny, T.; Brueggemann, E.E.; Ward, M.D.; Cazares, L.H.; Bavari, S. Sphingosine kinase 2 is a chikungunya virus host factor co-localized with the viral replication complex. Emerg. Microbes Infect. 2015, 4, e61. [Google Scholar] [CrossRef] [PubMed]
- Gorbalenya, A.E.; Koonin, E.V.; Lai, M.M. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett. 1991, 288, 201–205. [Google Scholar] [CrossRef]
- Koonin, E.V.; Gorbalenya, A.E.; Purdy, M.A.; Rozanov, M.N.; Reyes, G.R.; Bradley, D.W. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: Delineation of an additional group of positive-strand RNA plant and animal viruses. Proc. Natl. Acad. Sci. USA 1992, 89, 8259–8263. [Google Scholar] [CrossRef] [PubMed]
- Rack, J.G.; Perina, D.; Ahel, I. Macrodomains: Structure, Function, Evolution and Catalytic Activities. Annu. Rev. Biochem. 2016, 85, 431–454. [Google Scholar] [CrossRef] [PubMed]
- Lykouras, M.V.; Tsika, A.C.; Lichiere, J.; Papageorgiou, N.; Coutard, B.; Bentrop, D.; Spyroulias, G.A. NMR study of non-structural proteins-part III: (1)H, (13)C, (15)N backbone and side-chain resonance assignment of macro domain from Chikungunya virus (CHIKV). Biomol. NMR Assign. 2017. [Google Scholar] [CrossRef] [PubMed]
- Melekis, E.; Tsika, A.C.; Lichiere, J.; Chasapis, C.T.; Margiolaki, I.; Papageorgiou, N.; Coutard, B.; Bentrop, D.; Spyroulias, G.A. NMR study of non-structural proteins—Part I: (1)H, (13)C, (15)N backbone and side-chain resonance assignment of macro domain from Mayaro virus (MAYV). Biomol. NMR Assign. 2015, 9, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Makrynitsa, G.I.; Ntonti, D.; Marousis, K.D.; Tsika, A.C.; Lichiere, J.; Papageorgiou, N.; Coutard, B.; Bentrop, D.; Spyroulias, G.A. NMR study of non-structural proteins—Part II: (1)H, (13)C, (15)N backbone and side-chain resonance assignment of macro domain from Venezuelan equine encephalitis virus (VEEV). Biomol. NMR Assign. 2015, 9, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Malet, H.; Coutard, B.; Jamal, S.; Dutartre, H.; Papageorgiou, N.; Neuvonen, M.; Ahola, T.; Forrester, N.; Gould, E.A.; Lafitte, D.; et al. The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket. J. Virol. 2009, 83, 6534–6545. [Google Scholar] [CrossRef] [PubMed]
- Fehr, A.R.; Jankevicius, G.; Ahel, I.; Perlman, S. Viral Macrodomains: Unique Mediators of Viral Replication and Pathogenesis. Trends Microbiol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Neuvonen, M.; Ahola, T. Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites. J. Mol. Biol. 2009, 385, 212–225. [Google Scholar] [CrossRef] [PubMed]
- Egloff, M.P.; Malet, H.; Putics, A.; Heinonen, M.; Dutartre, H.; Frangeul, A.; Gruez, A.; Campanacci, V.; Cambillau, C.; Ziebuhr, J.; et al. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 2006, 80, 8493–8502. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Debing, Y.; Jankevicius, G.; Neyts, J.; Ahel, I.; Coutard, B.; Canard, B. Viral Macro Domains Reverse Protein ADP-Ribosylation. J. Virol. 2016, 90, 8478–8486. [Google Scholar] [CrossRef] [PubMed]
- Eckei, L.; Krieg, S.; Butepage, M.; Lehmann, A.; Gross, A.; Lippok, B.; Grimm, A.R.; Kummerer, B.M.; Rossetti, G.; Luscher, B.; et al. The conserved macrodomains of the non-structural proteins of Chikungunya virus and other pathogenic positive strand RNA viruses function as mono-ADP-ribosylhydrolases. Sci. Rep. 2017, 7, 41746. [Google Scholar] [CrossRef] [PubMed]
- McPherson, R.L.; Abraham, R.; Sreekumar, E.; Ong, S.E.; Cheng, S.J.; Baxter, V.K.; Kistemaker, H.A.; Filippov, D.V.; Griffin, D.E.; Leung, A.K. ADP-ribosylhydrolase activity of Chikungunya virus macrodomain is critical for virus replication and virulence. Proc. Natl. Acad. Sci. USA 2017, 114, 1666–1671. [Google Scholar] [CrossRef] [PubMed]
- Lulla, A.; Lulla, V.; Merits, A. Macromolecular assembly-driven processing of the 2/3 cleavage site in the alphavirus replicase polyprotein. J. Virol. 2012, 86, 553–565. [Google Scholar] [CrossRef] [PubMed]
- De, I.; Fata-Hartley, C.; Sawicki, S.G.; Sawicki, D.L. Functional analysis of nsP3 phosphoprotein mutants of Sindbis virus. J. Virol. 2003, 77, 13106–13116. [Google Scholar] [CrossRef] [PubMed]
- Atasheva, S.; Akhrymuk, M.; Frolova, E.I.; Frolov, I. New PARP gene with an anti-alphavirus function. J. Virol. 2012, 86, 8147–8160. [Google Scholar] [CrossRef] [PubMed]
- Shin, G.; Yost, S.A.; Miller, M.T.; Elrod, E.J.; Grakoui, A.; Marcotrigiano, J. Structural and functional insights into alphavirus polyprotein processing and pathogenesis. Proc. Natl. Acad. Sci. USA 2012, 109, 16534–16539. [Google Scholar] [CrossRef] [PubMed]
- Vihinen, H.; Saarinen, J. Phosphorylation site analysis of Semliki forest virus nonstructural protein 3. J. Biol. Chem. 2000, 275, 27775–27783. [Google Scholar] [CrossRef] [PubMed]
- Oberste, M.S.; Parker, M.D.; Smith, J.F. Complete sequence of Venezuelan equine encephalitis virus subtype IE reveals conserved and hypervariable domains within the C terminus of nsP3. Virology 1996, 219, 314–320. [Google Scholar] [CrossRef] [PubMed]
- Meissner, J.D.; Huang, C.Y.; Pfeffer, M.; Kinney, R.M. Sequencing of prototype viruses in the Venezuelan equine encephalitis antigenic complex. Virus Res. 1999, 64, 43–59. [Google Scholar] [CrossRef]
- Aaskov, J.; Jones, A.; Choi, W.; Lowry, K.; Stewart, E. Lineage replacement accompanying duplication and rapid fixation of an RNA element in the nsP3 gene in a species of alphavirus. Virology 2011, 410, 353–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, N.L.; Willis, L.V.; Smith, J.F.; Johnston, R.E. In vitro synthesis of infectious venezuelan equine encephalitis virus RNA from a cDNA clone: Analysis of a viable deletion mutant. Virology 1989, 171, 189–204. [Google Scholar] [CrossRef]
- Foy, N.J.; Akhrymuk, M.; Shustov, A.V.; Frolova, E.I.; Frolov, I. Hypervariable domain of nonstructural protein nsP3 of Venezuelan equine encephalitis virus determines cell-specific mode of virus replication. J. Virol. 2013, 87, 7569–7584. [Google Scholar] [CrossRef] [PubMed]
- Wootton, J.C.; Federhen, S. Analysis of compositionally biased regions in sequence databases. Methods Enzym. 1996, 266, 554–571. [Google Scholar]
- Dosztanyi, Z.; Csizmok, V.; Tompa, P.; Simon, I. IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 2005, 21, 3433–3434. [Google Scholar] [CrossRef] [PubMed]
- Galbraith, S.E.; Sheahan, B.J.; Atkins, G.J. Deletions in the hypervariable domain of the nsP3 gene attenuate Semliki Forest virus virulence. J. Gen. Virol. 2006, 87 Pt 4, 937–947. [Google Scholar] [CrossRef] [PubMed]
- Remenyi, R.; Roberts, G.C.; Zothner, C.; Merits, A.; Harris, M. SNAP-tagged Chikungunya Virus Replicons Improve Visualisation of Non-Structural Protein 3 by Fluorescence Microscopy. Sci. Rep. 2017, 7, 5682. [Google Scholar] [CrossRef] [PubMed]
- Schulte, T.; Liu, L.; Panas, M.D.; Thaa, B.; Dickson, N.; Gotte, B.; Achour, A.; McInerney, G.M. Combined structural, biochemical and cellular evidence demonstrates that both FGDF motifs in alphavirus nsP3 are required for efficient replication. Open Biol. 2016, 6, 160078. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Intrinsic disorder-based protein interactions and their modulators. Curr. Pharm. Des. 2013, 19, 4191–4213. [Google Scholar] [CrossRef] [PubMed]
- Nargi-Aizenman, J.L.; Simbulan-Rosenthal, C.M.; Kelly, T.A.; Smulson, M.E.; Griffin, D.E. Rapid activation of poly(ADP-ribose) polymerase contributes to Sindbis virus and staurosporine-induced apoptotic cell death. Virology 2002, 293, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Li, G.P.; La Starza, M.W.; Hardy, W.R.; Strauss, J.H.; Rice, C.M. Phosphorylation of Sindbis virus nsP3 in vivo and in vitro. Virology 1990, 179, 416–427. [Google Scholar] [CrossRef]
- Das, I.; Basantray, I.; Mamidi, P.; Nayak, T.K.; B, M.P.; Chattopadhyay, S.; Chattopadhyay, S. Heat shock protein 90 positively regulates Chikungunya virus replication by stabilizing viral non-structural protein nsP2 during infection. PLoS ONE 2014, 9, e100531. [Google Scholar] [CrossRef] [PubMed]
- Mohankumar, V.; Dhanushkodi, N.R.; Raju, R. Sindbis virus replication, is insensitive to rapamycin and torin1 and suppresses Akt/mTOR pathway late during infection in HEK cells. Biochem. Biophys. Res. Commun. 2011, 406, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.K.; Hardy, R.W. Role for the phosphatidylinositol 3-kinase-Akt-TOR pathway during sindbis virus replication in arthropods. J. Virol. 2012, 86, 3595–3604. [Google Scholar] [CrossRef] [PubMed]
- Thaa, B.; Biasiotto, R.; Eng, K.; Neuvonen, M.; Gotte, B.; Rheinemann, L.; Mutso, M.; Utt, A.; Varghese, F.; Balistreri, G.; et al. Differential Phosphatidylinositol-3-Kinase-Akt-mTOR Activation by Semliki Forest and Chikungunya Viruses Is Dependent on nsP3 and Connected to Replication Complex Internalization. J. Virol. 2015, 89, 11420–11437. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.K.; Liu, Q.; Tikoo, S.K.; Babiuk, L.A.; Zhou, Y. Influenza A virus NS1 protein activates the phosphatidylinositol 3-kinase (PI3K)/Akt pathway by direct interaction with the p85 subunit of PI3K. J. Gen. Virol. 2007, 88 Pt 1, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Strunk, U.; Saffran, H.A.; Wu, F.W.; Smiley, J.R. Role of herpes simplex virus VP11/12 tyrosine-based motifs in binding and activation of the Src family kinase Lck and recruitment of p85, Grb2 and Shc. J. Virol. 2013, 87, 11276–11286. [Google Scholar] [CrossRef] [PubMed]
- Songyang, Z.; Shoelson, S.E.; Chaudhuri, M.; Gish, G.; Pawson, T.; Haser, W.G.; King, F.; Roberts, T.; Ratnofsky, S.; Lechleider, R.J.; et al. SH2 domains recognize specific phosphopeptide sequences. Cell 1993, 72, 767–778. [Google Scholar] [PubMed]
- Saksela, K.; Permi, P. SH3 domain ligand binding: What’s the consensus and where’s the specificity? FEBS Lett. 2012, 586, 2609–2614. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, T.; Li, L.; Li, S.S. The SH3 domain—A family of versatile peptide- and protein-recognition module. Front. Biosci. 2008, 13, 4938–4952. [Google Scholar] [CrossRef] [PubMed]
- Zech, B.; Kurtenbach, A.; Krieger, N.; Strand, D.; Blencke, S.; Morbitzer, M.; Salassidis, K.; Cotten, M.; Wissing, J.; Obert, S.; et al. Identification and characterization of amphiphysin II as a novel cellular interaction partner of the hepatitis C virus NS5A protein. J. Gen. Virol. 2003, 84 Pt 3, 555–560. [Google Scholar] [CrossRef] [PubMed]
- Fros, J.J.; Domeradzka, N.E.; Baggen, J.; Geertsema, C.; Flipse, J.; Vlak, J.M.; Pijlman, G.P. Chikungunya virus nsP3 blocks stress granule assembly by recruitment of G3BP into cytoplasmic foci. J. Virol. 2012, 86, 10873–10879. [Google Scholar] [CrossRef] [PubMed]
- Tourriere, H.; Chebli, K.; Zekri, L.; Courselaud, B.; Blanchard, J.M.; Bertrand, E.; Tazi, J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 2003, 160, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.; Panas, M.D.; Achorn, C.A.; Lyons, S.; Tisdale, S.; Hickman, T.; Thomas, M.; Lieberman, J.; McInerney, G.M.; Ivanov, P.; et al. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 2016, 212, 845–860. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P.; Kedersha, N. Stress granules: The Tao of RNA triage. Trends Biochem. Sci. 2008, 33, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Buchan, J.R.; Parker, R. Eukaryotic stress granules: The ins and outs of translation. Mol. Cell 2009, 36, 932–941. [Google Scholar] [CrossRef] [PubMed]
- McInerney, G.M.; Kedersha, N.L.; Kaufman, R.J.; Anderson, P.; Liljestrom, P. Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus translation regulation. Mol. Biol. Cell 2005, 16, 3753–3763. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, O. Crystal structure of the G3BP2 NTF2-like domain in complex with a canonical FGDF motif peptide. Biochem. Biophys. Res. Commun. 2015, 467, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Vognsen, T.; Moller, I.R.; Kristensen, O. Crystal structures of the human G3BP1 NTF2-like domain visualize FxFG Nup repeat specificity. PLoS ONE 2013, 8, e80947. [Google Scholar] [CrossRef] [PubMed]
- Scholte, F.E.; Tas, A.; Albulescu, I.C.; Zusinaite, E.; Merits, A.; Snijder, E.J.; van Hemert, M.J. Stress granule components G3BP1 and G3BP2 play a proviral role early in Chikungunya virus replication. J. Virol. 2015, 89, 4457–4469. [Google Scholar] [CrossRef] [PubMed]
- Pazman, C.; Mayes, C.A.; Fanto, M.; Haynes, S.R.; Mlodzik, M. Rasputin, the Drosophila homologue of the RasGAP SH3 binding protein, functions in ras- and Rho-mediated signaling. Development 2000, 127, 1715–1725. [Google Scholar] [PubMed]
- Vognsen, T.; Kristensen, O. Crystal structure of the Rasputin NTF2-like domain from Drosophila melanogaster. Biochem. Biophys. Res. Commun. 2012, 420, 188–192. [Google Scholar] [CrossRef] [PubMed]


| Host Protein Interacting with nsP3 | Cell Type | Virus | Ref. |
|---|---|---|---|
| 14-3-3 z | C710 | SINV | [18] |
| 14-3-3 β/α | BHK-21 | SINV | [60] |
| 14-3-3 γ | BHK-21 | SINV | [60] |
| 14-3-3 ε | Rat2, HEK293 | SINV | [46] |
| BHK-21 | SINV | [60] | |
| C710 | SINV | [18] | |
| 14-3-3 ζ | Rat2, HEK293 | SINV | [46] |
| 14-3-3 ζ/δ | BHK-21 | SINV | [60] |
| 14-3-3 η | Rat2, HEK293 | SINV | [46] |
| 14-3-3 τ | BHK-21 | SINV | [60] |
| Amphiphysin-1 | In vitro, 293FT, N2A | SFV, SINV, CHIKV | [62] |
| Amphiphysin-2/BIN1—Bridging integrator 1 | In vitro | CHIKV, SFV | [69] |
| BHK-21 | SINV | [67] | |
| In vitro, 293FT, HeLa | SFV, SINV, CHIKV | [62] | |
| NIH 3T3 | CHIKV, SINV | [66] | |
| B23.2 | BHK-21 | SINV | [60] |
| Beta-actin | BHK-21 | SINV | [60] |
| Beta-tubulin | BHK-21 | SINV | [60] |
| CAPZA1—F-actin-capping protein subunit alpha-1 | BHK-21 | VEEV | [67] |
| NIH 3T3 | EEEV, VEEV | [66] | |
| CAPZA2—F-actin-capping protein subunit alpha-2 | NIH 3T3 | EEEV, VEEV | [66] |
| CAPZB—F-actin-capping protein subunit beta | BHK-21 | VEEV | [67] |
| NIH 3T3 | EEEV, VEEV | [66] | |
| CD2AP/CMS—CD2-associated protein | BHK-21 | VEEV | [67] |
| In vitro | SFV, SINV, CHIKV | [62] | |
| NIH 3T3 | EEEV, VEEV, CHIKV | [66] | |
| CKAP4—Cytoskeleton-associated protein 4 | Rat2 | SINV | [46] |
| CLTC—Clathrin heavy chain | NIH 3T3 | EEEV | [66] |
| DDX1—DEAD box polypeptide 1 | U87MG | VEEV | [70] |
| DDX3—DEAD/H box polypeptide 3 | U87MG | VEEV | [70] |
| DDX5—DEAD box polypeptide 5 | Rat2 | SINV | [46] |
| NIH 3T3 | SINV | [66] | |
| DDX6—DEAD box polypeptide 6 | NIH 3T3 | SINV | [66] |
| DDX17—DEAD box polypeptide 17 | Rat2 | SINV | [46] |
| NIH 3T3 | SINV | [66] | |
| Desmin | BHK-21 | SINV | [60] |
| DHX9—DEAD/H Box Polypeptide 9/ATP-dependent RNA helicase A | Rat2 | SINV | [46] |
| DNAJC9—DnaJ heat shock protein family (Hsp40) member C9 | NIH 3T3 | CHIKV, SINV | [66] |
| Dnmt1—DNA (cytosine-5-)-methyltransferase 1 | Rat2 | SINV | [46] |
| eEF1A—Eukaryotic translation elongation factor 1A | BHK-21 | SINV | [60] |
| eEF2—Eukaryotic translation elongation factor 2 | Rat2 | SINV | [46] |
| FHL2—Four and a half LIM domains protein 2 | NIH 3T3 | CHIKV | [66] |
| FMR1 | BHK-21 | VEEV | [67] |
| NIH 3T3 | EEEV, VEEV | [66] | |
| FXR1 | U87MG | VEEV | [70] |
| BHK-21 | VEEV | [67] | |
| BHK-21 | SINV | [60] | |
| NIH 3T3 | EEEV, VEEV | [66] | |
| FXR2 | BHK-21 | VEEV | [67] |
| NIH 3T3 | EEEV, VEEV | [66] | |
| G3BP-1/Rasputin | BHK-21 | SINV | [60] |
| Rat2, HEK293 | SINV | [46] | |
| BHK-21 | SINV | [67] | |
| BHK-21, MEF, HEK293 | SFV | [53] | |
| In vitro | SFV | [52] | |
| BHK-21 | SINV | [18] | |
| NIH 3T3 | EEEV, CHIKV, SINV | [66] | |
| Sf21 | CHIKV | [71] | |
| C710 | SINV | [18] | |
| G3BP-2 | BHK-21 | SINV | [67] |
| BHK-21, MEF | SFV | [53] | |
| BHK-21 | SINV | [18] | |
| NIH 3T3 | EEEV, CHIKV, SINV | [66] | |
| Rat2, HEK293 | SINV | [46] | |
| GRP78—78 kDa glucose-regulated protein | Rat2 | SINV | [46] |
| C710 | SINV | [18] | |
| GSTM1—Glutathione S-transferase Mu 1 | Rat2 | SINV | [46] |
| HELQ—ATP-dependent DNA helicase Hel308 | Rat2 | SINV | [46] |
| HIST1H1C—Histone cluster 1 H1 family member C | NIH 3T3 | CHIKV | [66] |
| hnRNP A0 | BHK-21 | SINV | [60] |
| hnRNP A1 | Rat2 | SINV | [46] |
| BHK-21 | SINV | [60] | |
| hnRNP A1-like | U87MG | VEEV | [70] |
| hnRNP A2/B1 | Rat2 | SINV | [46] |
| BHK-21 | SINV | [60] | |
| hnRNP A3 | Rat2 | SINV | [46] |
| BHK-21 | SINV | [60] | |
| hnRNP C | HeLa | SFV | [61] |
| hnRNP G | Rat2 | SINV | [46] |
| hnRNP K | HeLa | SFV | [61] |
| HeLa, BHK-21 | SINV | [72] | |
| hnRNP M | HeLa | SFV | [61] |
| hnRNP U | BHK-21 | SINV | [60] |
| HSC70—Heat shock cognate 71kDa protein | Rat2 | SINV | [46] |
| BHK-21 | SINV | [60] | |
| BHK-21, C710 | SINV | [18] | |
| HSP90—Heat shock protein 90 | 293T | CHIKV | [73] |
| HSPA1B—Heat shock protein 1B | NIH 3T3 | EEEV, VEEV, CHIKV, SINV | [66] |
| IGF2BP1—Insulin-like growth factor 2 mRNA-binding protein 1 | NIH 3T3 | EEEV, SINV | [66] |
| IGF2BP2 | NIH 3T3 | EEEV, SINV | [66] |
| IGF2BP3 | NIH 3T3 | EEEV, VEEV, CHIKV, SINV | [66] |
| IKKβ—Inhibitor of nuclear factor kappa-B kinase subunit beta | U87MG | VEEV, WEEV | [65] |
| INTS6—Integrator Complex Subunit 6/RNA helicase HDB/DICE1 | Rat2 | SINV | [46] |
| JUN—Jun proto-oncogene | NIH 3T3 | CHIKV | [66] |
| Lysophospholipase 1,2 | C710 | SINV | [18] |
| MAP 1B—Microtubule-associated protein 1B | U87MG | VEEV | [70] |
| MYBBP1A—MYB binding protein 1A | NIH 3T3 | CHIKV, SINV | [66] |
| MYH9—Myosin Heavy Chain 9 | BHK-21 | SINV | [60] |
| Myosin regulatory light chain | BHK-21 | SINV | [60] |
| NAP1L1—Nucleosome assembly protein 1-like 1 | NIH 3T3 | CHIKV | [66] |
| NAP1L4—Nucleosome assembly protein 1-like 4 | NIH 3T3 | CHIKV | [66] |
| Nucleolin | Rat2 | SINV | [46] |
| p85—PI3K subunit p85 | HEK293, BHK-21 | SFV, RRV | [63] |
| PARP-1—Poly(ADP-ribose) polymerase-1 | NSC34, 293T | SINV | [64] |
| PCBP1/hnRNP E1—Poly(rC)-binding protein 1 | HeLa | SFV | [61] |
| Peripherin 1 | Rat2 | SINV | [46] |
| PGAM5—PGAM family member 5, serine/threonine phosphatase | NIH 3T3 | EEEV, VEEV, CHIKV | [66] |
| Plectin 1 | BHK-21 | SINV | [60] |
| U87MG | VEEV | [70] | |
| PSF—PTB-associated splicing factor | Rat2 | SINV | [46] |
| RACK1/GNB2L1—Receptor of activated protein C kinase 1/Guanine nucleotide-binding protein subunit beta-2-like 1 | Rat2 | SINV | [46] |
| RDX—Radixin | NIH 3T3 | CHIKV | [66] |
| Ribosomal protein L10 | BHK-21 | SINV | [60] |
| Ribosomal protein L10A | U87MG | VEEV | [70] |
| BHK-21 | SINV | [60] | |
| C710 | SINV | [18] | |
| Ribosomal protein L15 | Rat2 | SINV | [46] |
| Ribosomal protein L18A | Rat2 | SINV | [46] |
| Ribosomal protein L23A | C710 | SINV | [18] |
| Ribosomal protein L3 | Rat2 | SINV | [46] |
| Ribosomal protein L4 | C710 | SINV | [18] |
| Ribosomal protein L5 | BHK-21 | SINV | [18] |
| Ribosomal protein L6 | U87MG | VEEV | [70] |
| BHK-21 | SINV | [18] | |
| Ribosomal protein L7 | BHK-21 | SINV | [60] |
| C710 | SINV | [18] | |
| Ribosomal protein L7A | BHK-21 | SINV | [18] |
| Ribosomal protein L8 | C710 | SINV | [18] |
| Ribosomal protein P0 | U87MG | VEEV | [70] |
| Ribosomal protein S10 | Rat2 | SINV | [46] |
| Ribosomal protein S18 | BHK-21 | SINV | [60] |
| Ribosomal protein S3 | Rat2 | SINV | [46] |
| Ribosomal protein S8 | U87MG | VEEV | [70] |
| Rat2 | SINV | [46] | |
| Ribosomal protein S9 | Rat2 | SINV | [46] |
| BHK-21 | SINV | [60] | |
| S100A4—S100 calcium binding protein A4 | NIH 3T3 | EEEV, VEEV | [66] |
| SASH1—SAM and SH3 domain-containing protein 1 | In vitro | SFV, SINV, CHIKV | [62] |
| SH3BP-5—SH3-domain binding protein 5 | Rat2 | SINV | [46] |
| SH3KBP1/CIN85—SH3 domain-containing kinase-binding protein 1 | In vitro | SFV, SINV, CHIKV | [62] |
| NIH 3T3 | EEEV, VEEV, CHIKV | [66] | |
| SK2—Sphingosine kinase 2 | HeLa | CHIKV | [74] |
| SLC25A13—Solute carrier family 25 member 13 | NIH 3T3 | EEEV, VEEV | [66] |
| SLC25A5—Solute carrier family 25 member 5/Adenine nucleotide translocator 2 | NIH 3T3 | EEEV, VEEV | [66] |
| Rat2 | SINV | [46] | |
| SNX33—Sorting nexin-33 | NIH 3T3 | EEEV | [66] |
| SNX9—Sorting nexin-9 | NIH 3T3 | EEEV | [66] |
| TRBP—TAR RNA-binding protein | C710 | SINV | [18] |
| Vimentin | BHK-21 | SINV | [60] |
| WDR48—WD repeat-containing protein 48 | BHK-21 | SINV | [67] |
| NIH 3T3 | SINV | [66] | |
| YBX1—Y-box-binding protein 1 | BHK-21 | SINV | [60] |
| BHK-21 | SINV | [18] | |
| NIH 3T3 | EEEV | [66] | |
| YBX3—Y-box-binding protein 3 | NIH 3T3 | CHIKV, SINV | [66] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Götte, B.; Liu, L.; McInerney, G.M. The Enigmatic Alphavirus Non-Structural Protein 3 (nsP3) Revealing Its Secrets at Last. Viruses 2018, 10, 105. https://doi.org/10.3390/v10030105
Götte B, Liu L, McInerney GM. The Enigmatic Alphavirus Non-Structural Protein 3 (nsP3) Revealing Its Secrets at Last. Viruses. 2018; 10(3):105. https://doi.org/10.3390/v10030105
Chicago/Turabian StyleGötte, Benjamin, Lifeng Liu, and Gerald M. McInerney. 2018. "The Enigmatic Alphavirus Non-Structural Protein 3 (nsP3) Revealing Its Secrets at Last" Viruses 10, no. 3: 105. https://doi.org/10.3390/v10030105
APA StyleGötte, B., Liu, L., & McInerney, G. M. (2018). The Enigmatic Alphavirus Non-Structural Protein 3 (nsP3) Revealing Its Secrets at Last. Viruses, 10(3), 105. https://doi.org/10.3390/v10030105