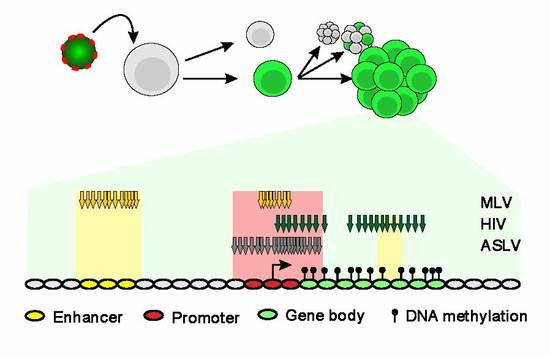
Proviruses with Long-Term Stable Expression Accumulate in Transcriptionally Active Chromatin Close to the Gene Regulatory Elements: Comparison of ASLV-, HIV- and MLV-Derived Vectors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Construction of the Retroviral Vectors
2.2. Cell Culture and Virus Propagation
2.3. Infection and Subcloning of K562 Cells
2.4. Cloning and Sequencing of Provirus Integration Sites
2.5. Mapping and Genomic Characterization of Provirus Integration Sites
2.6. Uniquely Mapped Matched Random Controls
2.7. Data Source and Integration Site Analysis
2.8. Active Genes
2.9. Merged Chromatin Segments
2.10. Active Gene Matched Random Controls
2.11. Statistics
3. Results
3.1. Stability of Proviral Expression
3.2. Gene Targeting and Distance to TSS
3.3. Stably Active Proviruses Associate with Active Genes
3.4. Stably Active Proviruses Associate with Active Chromatin and Active Regulatory Segments
3.5. Enhancer Proximity Is Not a Function of Active Gene Targeting
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Trinité, B.; Ohlson, E.C.; Voznesensky, I.; Rana, S.P.; Chan, C.N.; Mahajan, S.; Alster, J.; Burke, S.A.; Wodarz, D.; Levy, D.N. An HIV-1 replication pathway utilizing reverse transcription products that fail to integrate. J. Virol. 2013, 87, 12701–12720. [Google Scholar] [CrossRef] [PubMed]
- Elleder, D.; Pavlíček, A.; Pačes, J.; Hejnar, J. Preferential integration of human immunodeficiency virus type 1 into genes, cytogenetic R bands and GC-rich DNA regions: Insight from the human genome sequence. FEBS Lett. 2002, 517, 285–286. [Google Scholar] [CrossRef]
- Schröder, A.R.; Shinn, P.; Chen, H.; Berry, C.; Ecker, J.R.; Bushman, F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 2002, 110, 521–529. [Google Scholar] [CrossRef]
- Wu, X.; Li, Y.; Crise, B.; Burgess, S.M. Transcription start regions in the human genome are favored targets for MLV integration. Science 2003, 300, 1749–1751. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, R.S.; Beitzel1, B.F.; Schroder, A.R.W.; Shinn, P.; Chen, H.; Berry, C.C.; Ecker, J.R.; Bushman, J.D. Retroviral DNA integration: ASLV, HIV and MLV show distinct target site preferences. PLoS Biol. 2004, 2, e234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crise, B.; Li, Y.; Yuan, C.; Morcock, D.R.; Whitby, D.; Munroe, D.J.; Arthur, L.O.; Wu, X. Simian immunodeficiency virus integration preference is similar to that of human immunodeficiency virus type 1. J. Virol. 2005, 79, 12199–12204. [Google Scholar] [CrossRef] [PubMed]
- De Ravin, S.S.; Su, L.; Theobald, N.; Choi, U.; Macpherson, J.L.; Poidinger, M.; Symonds, G.; Pond, S.M.; Ferris, A.L.; Hughes, S.H.; et al. Enhancers are major targets for murine leukemia virus vector integration. J. Virol. 2014, 88, 4504–4513. [Google Scholar] [CrossRef] [PubMed]
- Narezkina, A.; Taganov, K.D.; Litwin, S.; Stoyanova, R.; Hayashi, J.; Seeger, C.; Skalka, A.M.; Katz, R.A. Genome-wide analyses of avian sarcoma virus integration sites. J. Virol. 2004, 78, 11656–11663. [Google Scholar] [CrossRef] [PubMed]
- Barr, S.D.; Leipzig, J.; Shinn, P.; Ecker, J.R.; Bushman, F.D. Integration targeting by avian sarcoma-leukosis virus and human immunodeficiency virus in the chicken genome. J. Virol. 2005, 79, 12035–12044. [Google Scholar] [CrossRef] [PubMed]
- Cherepanov, P.; Maertens, G.; Proost, P.; Devreese, B.; Van Beeumen, J.; Engelborghs, Y.; De Clercq, E.; Debyser, Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 2003, 278, 372–381. [Google Scholar] [CrossRef] [PubMed]
- Ciuffi, A.; Llano, M.; Poeschla, E.; Hoffmann, C.; Leipzig, J.; Shinn, P.; Ecker, J.R.; Bushman, F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 2005, 11, 1287–1289. [Google Scholar] [CrossRef] [PubMed]
- Vandekerckhove, L.; Christ, F.; Van Maele, B.; De Rijck, J.; Gijsbers, R.; Van den Haute, C.; Witvrouw, M.; Debyser, Z. Transient and stable knockdown of the integrase cofactor LEDGF/p75 reveals its role in the replication cycle of human immunodeficiency virus. J. Virol. 2006, 80, 1886–1896. [Google Scholar] [CrossRef] [PubMed]
- Marini, B.; Kertesz-Farkas, A.; Ali, H.; Lucic, B.; Lisek, K.; Manganaro, L.; Pongor, S.; Luzzati, R.; Recchia, A.; Mavilio, F.; et al. Nuclear architecture dictates HIV-1 integration site selection. Nature 2015, 521, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Sowd, G.A.; Serrao, E.; Wang, H.; Wang, W.; Fadel, H.J.; Poeschla, E.M.; Engelman, A.N. A critical role for alternative polyadenylation factor CPSF6 in targeting HIV-1 integration to transcriptionally active chromatin. Proc. Natl. Acad. Sci. USA 2016, 113, 1054–1063. [Google Scholar] [CrossRef] [PubMed]
- Rasheedi, S.; Shun, M.C.; Serrao, E.; Sowd, G.A.; Qian, J.; Hao, C.; Dasgupta, T.; Engelman, A.N.; Skowronski, J. The cleavage and polyadenylation specificity factor 6 (CPSF6) subunit of the capsid-recruited pre-messenger RNA cleavage factor I (CFIm) complex mediates HIV-1 integration into genes. J. Biol. Chem. 2016, 291, 11809–11819. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Larue, R.C.; Plumb, M.R.; Malani, N.; Male, F.; Slaughter, A.; Kessl, J.J.; Shkriabai, N.; Coward, E.; Aiyer, S.S.; et al. BET proteins promote efficient murine leukemia virus integration at transcription start sites. Proc. Natl. Acad. Sci. USA 2013, 110, 12036–12041. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.S.; Maetzig, T.; Maertens, G.N.; Sharif, A.; Rothe, M.; Weidner-Glunde, M.; Galla, M.; Schambach, A.; Cherepanov, P.; Schulz, T.F. Bromo- and extraterminal domain chromatin regulators serve as cofactors for murine leukemia virus integration. J. Virol. 2013, 87, 12721–12736. [Google Scholar] [CrossRef] [PubMed]
- De Rijck, J.; de Kogel, C.; Demeulemeester, J.; Vets, S.; El Ashkar, S.; Malani, N.; Bushman, F.D.; Landuyt, B.; Husson, S.J.; Busschots, K.; et al. The BET family of proteins targets moloney murine leukemia virus integration near transcription start sites. Cell Rep. 2013, 5, 886–894. [Google Scholar] [CrossRef] [PubMed]
- Winans, S.; Larue, R.C.; Abraham, C.M.; Shkriabai, N.; Skopp, A.; Winkler, D.; Kvaratskhelia, M.; Beemon, K.L. The FACT complex promotes avian leukosis virus DNA integration. J. Virol. 2017, 91, e00082-17. [Google Scholar] [CrossRef] [PubMed]
- Aiyer, S.; Swapna, G.V.; Malani, N.; Aramini, J.M.; Schneider, W.M.; Plumb, M.R.; Ghanem, M.; Larue, R.C.; Sharma, A.; Studamire, B.; et al. Altering murine leukemia virus integration through disruption of the integrase and BET protein family interaction. Nucleic Acids Res. 2014, 42, 5917–5928. [Google Scholar] [CrossRef] [PubMed]
- El Ashkar, S.; De Rijck, J.; Demeulemeester, J.; Vets, S.; Madlala, P.; Cermakova, K.; Debyser, Z.; Gijsbers, R. BET-independent MLV-based vectors target away from promoters and regulatory elements. Mol. Ther. Nucleic Acids 2014, 29, e179. [Google Scholar] [CrossRef] [PubMed]
- El Ashkar, S.; Van Looveren, D.; Schenk, F.; Vranckx, L.S.; Demeulemeester, J.; De Rijck, J.; Debyser, Z.; Modlich, U.; Gijsbers, R. Engineering next-generation BET-independent MLV vectors for safer gene therapy. Mol. Ther. Nucleic Acids 2017, 7, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Hacein-Bey-Abina, S.; Von Kalle, C.; Schmidt, M.; McCormack, M.P.; Wulffraat, N.; Leboulch, P.; Lim, A.; Osborne, C.S.; Pawliuk, R.; Morillon, E.; et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 2003, 302, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Schrijvers, R.; De Rijck, J.; Demeulemeester, J.; Adachi, N.; Vets, S.; Ronen, K.; Christ, F.; Bushman, F.D.; Debyser, Z.; Gijsbers, R. LEDGF/p75-independent HIV-1 replication demonstrates a role for HRP-2 and remains sensitive to inhibition by LEDGINs. PLoS Pathog. 2012, 8, e1002558. [Google Scholar] [CrossRef] [PubMed]
- Jordan, A.; Bisgrove, D.; Verdin, E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 2003, 22, 1868–1877. [Google Scholar] [CrossRef] [PubMed]
- Jordan, A.; Defechereux, P.; Verdin, E. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 2001, 20, 1726–1738. [Google Scholar] [CrossRef] [PubMed]
- Skupsky, R.; Burnett, J.C.; Foley, J.E.; Schaffer, D.V.; Arkin, A.P. HIV promoter integration site primarily modulates transcriptional burst size rather than frequency. PLoS Comput. Biol. 2010, 6, e1000952. [Google Scholar] [CrossRef] [PubMed]
- Lewinski, M.K.; Bisgrove, D.; Shinn, P.; Chen, H.; Hoffmann, C.; Hannenhalli, S.; Verdin, E.; Berry, C.C.; Ecker, J.R.; Bushman, F.D. Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J. Virol. 2005, 79, 6610–6619. [Google Scholar] [CrossRef] [PubMed]
- Blažková, J.; Trejbalová, K.; Gondois-Rey, F.; Halfon, P.; Philibert, P.; Guiguen, A.; Verdin, E.; Olive, D.; Van Lint, C.; Hejnar, J.; et al. CpG methylation controls reactivation of HIV from latency. PLoS Pathog. 2009, 5, e1000554. [Google Scholar] [CrossRef] [PubMed]
- Kauder, S.E.; Bosque, A.; Lindqvist, A.; Planelles, V.; Verdin, E. Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog. 2009, 5, e1000495. [Google Scholar] [CrossRef] [PubMed]
- Trejbalová, K.; Kovářová, D.; Blažková, J.; Machala, L.; Jilich, D.; Weber, J.; Kučerová, D.; Vencálek, O.; Hirsch, I.; Hejnar, J. Development of 5’LTR DNA methylation of latent HIV-1 provirus in cell line models and in long-term-infected individuals. Clin. Epigenet. 2016, 8, e19. [Google Scholar] [CrossRef] [PubMed]
- Sherrill-Mix, S.; Lewinski, M.K.; Famiglietti, M.; Bosque, A.; Malani, N.; Ocwieja, K.E.; Berry, C.C.; Looney, D.; Shan, L.; Agosto, L.M.; et al. HIV latency and integration site placement in five cell-based models. Retrovirology 2013, 10, e90. [Google Scholar] [CrossRef] [PubMed]
- Vranckx, L.S.; Demeulemeester, J.; Saleh, S.; Boll, A.; Vansant, G.; Schrijvers, R.; Weydert, C.; Battivelli, E.; Verdin, E.; Cereseto, A.; et al. LEDGIN-mediated Inhibition of Integrase-LEDGF/p75 Interaction Reduces Reactivation of Residual Latent HIV. EBioMedicine 2016, 8, 248–264. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.C.; Martinez, J.P.; Zorita, E.; Meyerhans, A.; Filion, G.J. Position effects influence HIV latency reversal. Nat. Struct. Mol. Biol. 2017, 24, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Svoboda, J.; Hejnar, J.; Geryk, J.; Elleder, D.; Vernerová, Z. Retroviruses in foreign species and the problem of provirus silencing. Gene 2000, 261, 181–188. [Google Scholar] [CrossRef]
- Lounková, A.; Dráberová, E.; Šenigl, F.; Trejbalová, K.; Geryk, J.; Hejnar, J.; Svoboda, J. Molecular events accompanying rous sarcoma virus rescue from rodent cells and the role of viral gene complementation. J. Virol. 2014, 88, 3505–3515. [Google Scholar] [CrossRef] [PubMed]
- Hejnar, J.; Svoboda, J.; Geryk, J.; Fincham, V.J.; Hák, R. High rate of morphological reversion in tumor cell line H-19 associated with permanent transcriptional suppression of the LTR, V-SRC, LTR provirus. Cell Growth Differ. 1994, 5, 277–285. [Google Scholar] [PubMed]
- Hejnar, J.; Plachý, J.; Geryk, J.; Machon, O.; Trejbalová, K.; Guntaka, R.V.; Svoboda, J. Inhibition of the rous sarcoma virus long terminal repeat-driven transcription by in vitro methylation: Different sensitivity in permissive chicken cells versus mammalian cells. Virology 1999, 255, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Šenigl, F.; Auxt, M.; Hejnar, J. Transcriptional provirus silencing as a crosstalk of de novo DNA methylation and epigenomic features at the integration site. Nucleic Acids Res. 2012, 40, 5298–5312. [Google Scholar] [CrossRef] [PubMed]
- Šenigl, F.; Miklík, D.; Auxt, M.; Hejnar, J. Accumulation of long-term transcriptionally active integrated retroviral vectors in active promoters and enhancers. Nucleic Acids Res. 2017, 45, 12752–12765. [Google Scholar] [CrossRef] [PubMed]
- Šenigl, F.; Plachý, J.; Hejnar, J. The core element of a CpG island protects avian sarcoma and leukosis virus-derived vectors from transcriptional silencing. J. Virol. 2008, 82, 7818–7827. [Google Scholar] [CrossRef] [PubMed]
- Kalina, J.; Šenigl, F.; Mičáková, A.; Mucksová, J.; Blažková, J.; Yan, H.; Poplštein, M.; Hejnar, J.; Trefil, P. Retrovirus-mediated in vitro gene transfer into chicken male germ line cells. Reproduction 2007, 134, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Uren, A.G.; Mikkers, H.; Kool, J.; van der Weyden, L.; Lund, A.H.; Wilson, C.H.; Rance, R.; Jonkers, J.; van Lohuizen, M.; Berns, A.; et al. A high-throughput splinkerette-PCR method for the isolation and sequencing of retroviral insertion sites. Nat. Protoc. 2009, 4, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Berry, C.C. Restrsiteutils: Restriction Site Distances and Matched Samples. R Package Version 1.2.8. 2017. Available online: http://www.bioconductor.org/packages/2.12/bioc/html/Restrsiteutils.html (accessed on 28 January 2018).
- Pagès, H.; Aboyoun, P.; Gentleman, R.; DebRoy, S. Biostrings: Efficient Manipulation of Biological Strings. R Package Version 2.46.0. 2017. Available online: http://www.bioconductor.org/packages/2.12/bioc/html/Biostrings.html (accessed on 28 January 2018).
- Lawrence, M.; Huber, W.; Pagès, H.; Aboyoun, P.; Carlson, M.; Gentleman, R.; Morgan, M.T.; Carey, V.J. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013, 9, e1003118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ernst, J.; Kellis, M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol. 2010, 28, 817–825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ernst, J.; Kellis, M. ChromHMM: Automating chromatin-state discovery and characterization. Nat. Methods 2012, 9, 215–216. [Google Scholar] [CrossRef] [PubMed]
- Kanamori-Katayama, M.; Itoh, M.; Kawaji, H.; Lassmann, T.; Katayama, S.; Kojima, M.; Bertin, N.; Kaiho, A.; Ninomiya, N.; Daub, C.O.; et al. Unamplified cap analysis of gene expression on a single-molecule sequencer. Genome Res. 2011, 21, 1150–1159. [Google Scholar] [CrossRef] [PubMed]
- Lusic, M.; Siliciano, R.F. Nuclear landscape of HIV-1 infection and integration. Nat. Rev. Microbiol. 2017, 15, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Hughes, S.H.; Coffin, J.M. What Integration Sites Tell Us about HIV Persistence. Cell Host Microbe 2016, 19, 588–598. [Google Scholar] [CrossRef] [PubMed]
- Mok, H.P.; Lever, A.M. Chromatin, gene silencing and HIV latency. Genome Biol. 2007, 8, e228. [Google Scholar] [CrossRef] [PubMed]
- LaFave, M.C.; Varshney, G.K.; Gildea, D.E.; Wolfsberg, T.G.; Baxevanis, A.D.; Burgess, S.M. MLV integration site selection is driven by strong enhancers and active promoters. Nucleic Acids Res. 2014, 42, 4257–4269. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, S.; Winans, S.; Lam, G.; Justice, J.; Morgan, R.; Beemon, K. Selection for avian leukosis virus integration sites determines the clonal progression of B-cell lymphomas. PLoS Pathog. 2017, 13, e1006708. [Google Scholar] [CrossRef] [PubMed]
- Justice, J.F.; Morgan, R.W.; Beemon, K.L. Common Viral Integration Sites Identified in Avian Leukosis Virus-Induced B-Cell Lymphomas. MBio 2015, 6, e01863. [Google Scholar] [CrossRef] [PubMed]
- Pajer, P.; Pečenka, V.; Králová, J.; Karafiát, V.; Průková, D.; Zemanová, Z.; Kodet, R.; Dvořák, M. Identification of potential human oncogenes by mapping the common viral integration sites in avian nephroblastoma. Cancer Res. 2006, 66, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Plachý, J.; Kotáb, J.; Divina, P.; Reinišová, M.; Šenigl, F.; Hejnar, J. Proviruses selected for high and stable expression of transduced genes accumulate in broadly transcribed genome areas. J. Virol. 2010, 84, 4204–4211. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.; Hellmann, I.; Stadler, M.B.; Ramos, L.; Paabo, S.; Rebhan, M.; Schübeler, D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007, 39, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Hodges, E.; Smith, A.D.; Kendall, J.; Xuan, Z.; Ravi, K.; Rooks, M.; Zhang, M.Q.; Ye, K.; Bhattacharjee, A.; Brizuela, L.; et al. High definition profiling of mammalian DNA methylation by array capture and single molecule bisulfite sequencing. Genome Res. 2009, 19, 1593–1605. [Google Scholar] [CrossRef] [PubMed]







© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miklík, D.; Šenigl, F.; Hejnar, J. Proviruses with Long-Term Stable Expression Accumulate in Transcriptionally Active Chromatin Close to the Gene Regulatory Elements: Comparison of ASLV-, HIV- and MLV-Derived Vectors. Viruses 2018, 10, 116. https://doi.org/10.3390/v10030116
Miklík D, Šenigl F, Hejnar J. Proviruses with Long-Term Stable Expression Accumulate in Transcriptionally Active Chromatin Close to the Gene Regulatory Elements: Comparison of ASLV-, HIV- and MLV-Derived Vectors. Viruses. 2018; 10(3):116. https://doi.org/10.3390/v10030116
Chicago/Turabian StyleMiklík, Dalibor, Filip Šenigl, and Jiří Hejnar. 2018. "Proviruses with Long-Term Stable Expression Accumulate in Transcriptionally Active Chromatin Close to the Gene Regulatory Elements: Comparison of ASLV-, HIV- and MLV-Derived Vectors" Viruses 10, no. 3: 116. https://doi.org/10.3390/v10030116
APA StyleMiklík, D., Šenigl, F., & Hejnar, J. (2018). Proviruses with Long-Term Stable Expression Accumulate in Transcriptionally Active Chromatin Close to the Gene Regulatory Elements: Comparison of ASLV-, HIV- and MLV-Derived Vectors. Viruses, 10(3), 116. https://doi.org/10.3390/v10030116