Phage Therapy: What Have We Learned?
Abstract
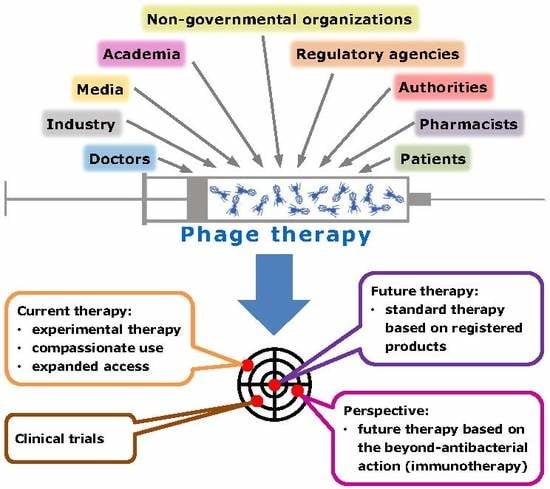
:1. More Room for Phage Therapy on the Horizon?
2. Doctors, Pharmacists, and Academia
3. CRISPR-Cas: From Phages to Eukaryotes
4. Patients, the Media, and PT
5. Industry and SMEs
6. Authorities and PT
7. National Regulations Enabling Experimental Therapy (Including PT)
8. Important Issues Which Need Addressing to Enable Further Progress and Optimization of PT and Relevant Clinical Trials
9. PT and Antibody Responses against Phages
10. Monotherapy vs. Phage Cocktails
11. Optimal Clinical Models for PT and Prognosis of Therapy
12. Mouse Model of Acute Urinary Tract Infection Confirms Neutrophil–Phage Synergy
13. Prophages in Bacterial Strains Used for Therapeutic Phage Propagation: Their Significance, Detection, and Elimination
Bacterial Strains for the Propagation of Therapeutic Phages
14. Prophage Detection Methods
15. Elimination of Prophages from Phage Propagation Strains
16. Future Possibilities to Produce Industrial Phage Propagation Strains
17. Surrogate Hosts for the Propagation of Therapeutic Phages
18. Economic Aspects of the Industrial Construction of Phage Propagation Strains
19. PT: Beyond the Antibacterial Action
20. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Borysowski, J.; Międzybrodzki, R.; Górski, A. Phage Therapy: Current Research and Application; Caister Academic Press: Norfolk, UK, 2014. [Google Scholar]
- Azeredo, J.; Sillankorva, J. (Eds.) Bacteriophage Therapy: From Lab to Clinical Practice; Springer Nature, Humana Press: New York, NY, USA, 2018; ISBN 978-1-4939-7395-8. [Google Scholar]
- Alvarez, D.R.; Abedon, S.T. An online phage therapy bibliography: Separating under-indexed wheat from overly indexed chaff. AIMS Microbiol. 2017, 3, 525–528. [Google Scholar]
- P.H.A.G.E. Phages for Human Applications Europe Group. Available online: www.p-h-a-g-e.org (accessed on 5 April 2018).
- Sybesma, W.; Rohde, C.; Bardy, P.; Pirnay, J.P.; Cooper, I.; Caplin, J.; Chanishvili, N.; Coffey, A.; De Vos, D.; Scholz, A.H.; et al. Silk route to the acceptance and re-implementation of bacteriophage therapy. Biotechnol. J. 2016, 11, 595–600. [Google Scholar] [CrossRef]
- Sybesma, W.; Rohde, C.; Bardy, P.; Pirnay, J.P.; Cooper, I.; Caplin, J.; Chanishvili, N.; Coffey, A.; De Vos, D.; Scholz, A.H.; et al. Silk Route to the Acceptance and Re-Implementation of Bacteriophage Therapy-Part II. Antibiotics 2018, 7, 35. [Google Scholar] [CrossRef]
- O’Reilly, C.E.; Jaron, P.; Ochieng, B.; Nyaguara, A.; Tate, J.E.; Parsons, M.B.; Bopp, C.A.; Williams, K.A.; Vinje, J.; Blanton, E.; et al. Risk factors for death among children less than 5 years old hospitalized with diarrhea in rural western Kenya, 2005–2007: A cohort study. PLoS Med. 2012, 9, e1001256. [Google Scholar] [CrossRef] [PubMed]
- Nagel, T.E.; Chan, B.K.; De Vos, D.; El-Shibiny, A.; Kang’ethe, E.; Makumi, A.; Pirnay, J.P. The developing world urgently needs phages to combat pathogenic bacteria. Front. Microbiol. 2016, 7, 882. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.phagesforglobalhealth.org/ (accessed on 24 May 2018).
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011, 471, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011, 9, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Hertsenberg, A.; AVROTROS TV. Dokters van Morgen over bacteriën (Update of emission of 21-03-2017 Bacteriofagen: een alternatief voor antibiotica?). Emission 24-10-2017. Available online: https://zorgnu.avrotros.nl/uitzending/24-10-2017/ (accessed on 24 May 2018).
- Das Erste. Phagen—hilfreiche Viren gegen bakterielle Infektionen. Available online: www.daserste.de/information/wissen-kultur/w-wie-wissen/videos/phagen-hilfreiche-viren-gegen-bakterielle-infektionen-100.html (accessed on 24 May 2018).
- Pranz, S.; Weiss, F. PhagenWagen. Der Spiegel Wissen 2017, 6, 64–69. [Google Scholar]
- Van Zonneveld, B. Getreuzel met de faag. Elsevier Weekblad 2017, 45, 70–71. [Google Scholar]
- Melchior, M. Update antibiotica. Kunnen we in de toekomstzonder? Libelle 2018, 6, 58–61. [Google Scholar]
- Pirnay, J.P.; Blasdel, B.G.; Bretaudeau, L.; Buckling, A.; Chanishvili, N.; Clark, J.R.; Corte-Real, S.; Debarbieux, L.; Dublanchet, A.; De Vos, D.; et al. Quality and safety requirements for sustainable phage therapy products. Pharm. Res. 2015, 32, 2173–2179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- BioIndustry-ein Service-Verbund mit internationalem Anspruch. Available online: https://www.bioindustry.de/nc/mitglieder/details.html?mtg=38 (accessed on 24 May 2018).
- European Commission. Action plan against the rising threats from antimicrobial resistance. COM (2011)748 and Road Map (Updated 16/11/2016). Available online: https://ec.europa.eu/health/amr/ (accessed on 24 May 2018).
- World Health Organization (WHO). Global action plan on antimicrobial resistance (document WHA68/2015/REC/1, Annex 3). Available online: http://apps.who.int/gb/ebwha/pdf_files/WHA68-REC1/A68_R1_REC1-en.pdf (accessed on 24 May 2018).
- The Food and Drug Administration (FDA). Bacteriophage Therapy: Scientific and Regulatory Issues Public Workshop. Available online: https://www.fda.gov/BiologicsBloodVaccines/NewsEvents/WorkshopsMeetingsConferences/ucm544294.htm (accessed on 24 May 2018).
- Schooley, R.T.; Biswas, B.; Gill, J.J.; Hernandez-Morales, A.; Lancaster, J.; Lessor, L.; Bar, J.J.; Reed, S.L.; Rohwer, F.; Benler, S.; et al. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumanniiinfection. Antimicrob. Agents Chemother. 2017, 61, e00954-17. [Google Scholar] [CrossRef] [PubMed]
- Intralytix Safety by Nature. Intralytix Receives FDA Clearance to Initiate Phase I/IIa Clinical Trials. Available online: http://www.intralytics.com (accessed on 30 March 2018).
- PhagoBurn Project Funded by the European Union under the 7th Framework Programme for Research and Development. Available online: www.phagoburn.eu (accessed on 4 May 2018).
- The White House. National Action Plan for Combating Antibiotic Resistant Bacteria. The White House Washington. March 2015, p. 44. Available online: https://obamawhitehouse.archives.gov/sites/default/files/docs/national_action_plan_for_combating_antibotic-resistant_bacteria.pdf (accessed on 24 May 2018).
- Transatlantic Taskforce Antimicrobial Resistance (TATFAR). Actions and Recommendations, Action 3.6. Available online: https://www.cdc.gov/drugresistance/tatfar/tatfar-recomendations.html (accessed on 24 May 2018).
- Federal Government. Deutsche Antimicrobiale Resistance Strategie DART 2020; Fighting Antibiotic Resistance for the Good of Both Humans and Animal; Decision by the Federal Cabinet of 13th of May 2015; Federal Government: Berlin, Germany, 2015; p. 24.
- Federal Government Germany. CombatingAntimicrobial Resistance; Examples of Best-Practices of the G7 Countries; Report of the G7 Meeting; Federal Government Germany: Berlin, Germany, 2015; p. 99.
- Pirnay, J.P.; Verbeken, G.; Ceyssens, P.J.; Huys, I.; De Vos, D.; Ameloot, C.; Fauconnier, A. The Magistral Phage. Viruses 2018, 10, 64. [Google Scholar] [CrossRef] [PubMed]
- World Medical Association (WMA). Declaration of Helsinki—Ethical Principles for Medicalresearch Involving Human Subjects; Article 37. Unproven interventions in clinical practice. Adopted by the 18th WMA General Assembly, Helsinki, Finland, June 1964 and Current Version as Amended 64th WMA General Assembly; WMA: Fortaleza, Brazil, 2013. [Google Scholar]
- Międzybrodzki, R.; Borysowski, J.; Weber-Dąbrowska, B.; Fortuna, W.; Letkiewicz, S.; Szufnarowski, K.; Pawełczyk, Z.; Rogóż, P.; Kłak, M.; Wojtasik, E.; et al. Clinical aspects of phage therapy. Adv. Virus Res. 2012, 83, 73–121. [Google Scholar] [CrossRef] [PubMed]
- Górski, A.; Międzybrodzki, R.; Weber-Dąbrowska, B.; Fortuna, W.; Letkiewicz, S.; Rogóż, P.; Jończyk-Matysiak, E.; Dąbrowska, K.; Majewska, J.; Borysowski, J. Phage therapy: Present and future. Front. Microbiol. 2016, 7, 1515. [Google Scholar] [CrossRef] [PubMed]
- European Commission Directorate-General for Health and Food Safety. Health Systems, Medical Products and Innovation. Medicines: Policy, Authorisation and Monitoring. STAMP Commission Expert Group. STAMP 4/22, Subject: Compassionate Use Programmes Agenda Item 6. 13 March 2016. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/Regulatory_and_procedural_guideline/2009/10/WC500004075.pdf (accessed on 4 May 2018).
- Balasubramanian, G.; Morampudi, S.; Chhabra, P.; Gowda, A.; Zomorodi, B. An overview of Compassionate Use Programs in the European Union member states. Intractable Rare Dis. Res. 2016, 5, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Directive 2001/83/EC of the European Parliament and of the Council. Available online: https://ec.europa.eu/health/sites/health/files/files/eudralex/vol-1/dir_2001_83_consol_2012/dir_2001_83_cons_2012_en.pdf (accessed on 24 May 2018).
- US Food and Drug Administration FDA. Expanded Access (Compassionate Use). Available online: https://www.fda.gov/NewsEvents/PublicHealthFocus/ExpandedAccessCompassionateUse/default.htm (accessed on 27 April 2018).
- US Food and Drug Administration FDA. Expanded Access to Investigational Drugs for Treatment Use. Questions and Answers. Guidance for Industry. June 2016; updated October 2017. Available online: https://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM351261.pdf (accessed on 4 May 2018).
- Joffe, S.; Lynch, H.F. Federal right-to-try legislation—Threatening the FDA’s public health mission. N. Engl. J. Med. 2018, 378, 695–697. [Google Scholar] [CrossRef] [PubMed]
- House of Representatives, 115th CONGRESS 2d Session. H.R. 5247. To authorize the use of eligible investigational drugs by eligible patients who have been diagnosed with a stage of a disease or condition in which there is reasonable likelihood that death will occur within a matter of months, or with another eligible illness, and for other purposes. Available online: https://www.congress.gov/bill/115th-congress/house-bill/5247/text/eh (accessed on 24 May 2018).
- Thomas, K. Why can’t dying patients get the drugs they want? NYT, 28 March 2018. [Google Scholar]
- Available online: https://www.canada.ca/content/dam/hc-sc/migration/hc-sc/dhp-mps/alt_formats/hpfb-dgpsa/pdf/acces/sapg3_pasg3-eng.pdf (accessed on 27 March 2018).
- Australian Government, Department of Health. Therapeutic Goods Administration. Accessing Unapproved Products. Available online: https://www.tga.gov.au/accessing-unapproved-products (accessed on 4 May 2018).
- Califf, R.M.; Ostroff, S. FDA as a catalyst for translation. Sci. Transl. Med. 2015, 7, 296ed9. [Google Scholar] [CrossRef] [PubMed]
- Międzybrodzki, R.; Hoyle, N.; Zhvaniya, F.; Gogokhia, L. Current updates from the long-standing phage research centers in Georgia, Poland, and Russia. In Bacteriophages; Harper, D., Abedon, S., Burrowes, B., McConville, M., Eds.; Springer: Cham, Switzerland, 2018; ISBN 978-3-319-40598-8. [Google Scholar]
- Majewska, J.; Beta, W.; Lecion, D.; Hodyra-Stefaniak, K.; Kłopot, A.; Kaźmierczak, Z.; Miernikiewicz, P.; Piotrowicz, A.; Ciekot, J.; Owczarek, B.; et al. Oral Application of T4 Phage Induces Weak Antibody Production in the Gut and in the Blood. Viruses 2015, 7, 4783–4799. [Google Scholar] [CrossRef] [PubMed]
- Łusiak-Szelachowska, M.; Żaczek, M.; Weber-Dąbrowska, M.; Międzybrodzki, R.; Letkiewicz, S.; Fortuna, W.; Rogóż, P.; Szufnarowski, K.; Jończyk-Matysiak, E.; Olchawa, E.; et al. Antiphage Activity of Sera in Patients During Phage Therapy in Relation to its Outcome. Future Microbiol. 2017, 12, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Persn com Międzybrodzki, R.; Rogóż, P.; Fortuna, W.; Wójcik, E.; Letkiewicz, A.; Weber-Dąbrowska, B.; Górski, A. Wrocław, Poland. The first retrospective analysis of long term results of the application of phage preparations in patients with chronic bacterial infections. Viruses Microbes 2018, in press. [Google Scholar]
- Brüssow, H. Special Issue Information. Available online: www.mdpi.com/journal/viruses/special_issues/Phagetherapy (accessed on 10 May 2018).
- Letkiewicz, S.; Miedzybrodzki, R.; Fortuna, W.; Weber-Dabrowska, B.; Górski, A. Eradication of Enterococcus faecalis by phage therapy in chronic bacterial prostatitis—Case report. Folia Microbiol. 2009, 54, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, S.; Baker, K.; Padman, B.S.; Patwa, R.; Dunstan, R.A.; Weston, T.A.; Schlosser, K.; Bailey, B.; Lithgow, T.; Lazarou, M.; et al. Bacteriophage transcytosis provides a mechanism to cross epithelial cell layers. mBio 2017, 8, e01874-17. [Google Scholar] [CrossRef] [PubMed]
- Lehti, T.A.; Pajunen, M.I.; Skog, M.S.; Finne, J. Internalization of a polysialic acid-binding Escherichia coli bacteriophage into eukaryotic neuroblastoma cells. Nat. Commun. 2017, 8, 1915. [Google Scholar] [CrossRef] [PubMed]
- Górski, A.; Międzybrodzki, R.; Borysowski, J.; Dąbrowska, K.; Wierzbicki, P.; Ohams, M.; Korczak-Kowalska, G.; Olszowska-Zaremba, N.; Łusiak-Szelachowska, M.; Kłak, M.; et al. Phage as a modulator of immune responses: Practical implications for phage therapy. Adv. Virus Res. 2012, 83, 41–71. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, W.J. Mouse model of ascending urinary tract infection. In Handbook of Animal Models of Infection; Zak, O., Sande, M.A., Eds.; Academic Press: London, UK, 1999; pp. 435–439. ISBN 0-12-776390-7. [Google Scholar]
- Kruisbeck, A.M. Isolation and fractionation of mononuclear cell populations. In Current Protocols in Immunology; Coligan, J.E., Kruisbeck, A.M., Margulies, D.H., Shevach, E.M., Strober, W., Eds.; Wiley: New York, NY, USA, 2000; pp. 3.1.1–3.1.5. [Google Scholar]
- Buisman, H.P.; Buys, L.F.M.; Langermans, J.A.M.; van den Broek, P.J.; van Furth, R. Effect of probenecid on phagocytosis and intracellular killing of Staphylococcus aureus and Escherichia coli by human monocytes and granulocytes. Immunology 1991, 74, 338–341. [Google Scholar] [PubMed]
- Leijh, P.C.J.; Van Zwet, T.L.; Van Furth, R. Effect of concanavalin A on intracellular killing of Staphylococcus aureus by human phagocytes. Clin. Exp. Immunol. 1984, 58, 557–565. [Google Scholar] [PubMed]
- Jończyk-Matysiak, E. The Effect of Bacteriophage Preparations on Intracellular Killing of Bacteria by Phagocytes. Ph.D. Thesis, Ludwik Hirszfeld Institute of Immunology and Experimental Therapy Polish Academy of Sciences, Wrocław, Poland, 2015; pp. 61–118. [Google Scholar]
- Roach, D.R.; Leung, C.Y.; Henry, M.; Morello, E.; Singh, D.; Di Santo, J.P.; Weitz, J.S.; Debarbieux, L. Synergy between the Host Immune System and Bacteriophage Is Essential for Successful Phage Therapy against an Acute Respiratory Pathogen. Cell Host Microbe 2017, 22, 38.e4–47.e4. [Google Scholar] [CrossRef] [PubMed]
- Łobocka, M.; Hejnowicz, M.S.; Gągała, U.; Weber-Dąbrowska, B.; Węgrzyn, G.; Dadlez, M. The first step to bacteriophage therapy—How to choose the correct phage. In Phage Therapy: Current Research and Applications; Borysowski, J., Międzybrodzki, R., Górski, A., Eds.; Caister Academic Press: Norfolk, UK, 2014; pp. 23–69. [Google Scholar]
- Rohde, C.; Resch, G.; Pirnay, J.P.; Blasdel, B.G.; Debarbieux, L.; Gelman, D.; Górski, A.; Hazan, R.; Huys, I.; Kakabadze, E.; et al. Expert Opinion on Three Phage Therapy Related Topics: Bacterial Phage Resistance, Phage Training and Prophages in Bacterial Production Strains. Viruses 2018, 10, E178. [Google Scholar] [CrossRef] [PubMed]
- Łobocka, M.; Hejnowicz, M.S.; Dąbrowski, K.; Izak, D.; Gozdek, A.; Głowacka, A.; Gawor, J.; Kosakowski, J.; Gromadka, R.; Weber-Dąbrowska, B.; et al. Staphylococcus aureus Strains for the Production of Monoclonal Bacteriophage Preparations Deprived of Contamination with Plasmid DNA. U.S. Patent WO 2016/030871 A1, 16 March 2016. [Google Scholar]
- Brussow, H.; Canchaya, C.; Hardt, W.D. Phages and the evolution of bacterial pathogens: From genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 2004, 68, 560–602. [Google Scholar] [CrossRef] [PubMed]
- Bossi, L.; Fuentes, J.A.; Mora, G.; Figueroa-Bossi, N. Prophage contribution to bacterial population dynamics. J. Bacteriol. 2003, 185, 6467–6471. [Google Scholar] [CrossRef] [PubMed]
- Fortier, L.C.; Sekulovic, O. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence 2013, 5, 354–365. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T.; Lejeune, J.T. Why bacteriophage encode exotoxins and other virulence factors. Evol. Bioinform. Online 2005, 1, 97–110. [Google Scholar] [CrossRef]
- Varani, A.M.; Monteiro-Vitorello, C.B.; Nakaya, H.I.; Van Sluys, M.A. The role of prophage in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 2013, 51, 429–451. [Google Scholar] [CrossRef] [PubMed]
- Davies, E.V.; Winstanley, C.; Fothergill, J.L.; James, C.E. The role of temperate bacteriophages in bacterial infection. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef] [PubMed]
- Trost, E.; Blom, J.; Soares Sde, C.; Huang, I.H.; Al-Dilaimi, A.; Schröder, J.; Jaenicke, S.; Dorella, F.A.; Rocha, F.S.; Miyoshi, A.; et al. Pangenomic study of Corynebacterium diphtheriae that provides insights into the genomic diversity of pathogenic isolates from cases of classical diphtheria, endocarditis, and pneumonia. J. Bacteriol. 2012, 194, 3199–3215. [Google Scholar] [CrossRef] [PubMed]
- The, H.C.; Thanh, D.P.; Holt, K.E.; Thomson, N.R.; Baker, S. The genomic signatures of Shigella evolution, adaptation and geographical spread. Nat. Rev. Microbiol. 2016, 4, 235–250. [Google Scholar] [CrossRef] [PubMed]
- Gyles, C.; Boerlin, P. Horizontally transferred genetic elements and their role in pathogenesis of bacterial disease. Vet. Pathol. 2014, 51, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Mai-Prochnow, A.; Hui, J.G.; Kjelleberg, S.; Rakonjac, J.; McDougald, D.; Rice, S.A. Big things in small packages: The genetics of filamentous phage and effects on fitness of their host. FEMS Microbiol. Rev. 2015, 39, 465–487. [Google Scholar] [CrossRef] [PubMed]
- Hare, J.M.; Ferrell, J.C.; Witkowski, T.A.; Grice, A.N. Prophage induction and differential RecA and UmuDAb transcriptome regulation in the DNA damage responses of Acinetobacter baumannii and Acinetobacter baylyi. PLoS ONE 2014, 9, e93861. [Google Scholar] [CrossRef] [PubMed]
- Repizo, G.D.; Viale, A.M.; Borges, V.; Cameranesi, M.M.; Taib, N.; Espariz, M.; Brochier-Armanet, C.; Gomes, J.P.; Salcedo, S.P. The Environmental Acinetobacter baumannii Isolate DSM30011 Reveals Clues into the Preantibiotic Era Genome Diversity, Virulence Potential, and Niche Range of a Predominant Nosocomial Pathogen. Genome Biol. Evol. 2017, 9, 2292–2307. [Google Scholar] [CrossRef] [PubMed]
- Touchon, M.; Cury, J.; Yoon, E.J.; Krizova, L.; Cerqueira, G.C.; Murphy, C.; Feldgarden, M.; Wortman, J.; Clermont, D.; Lambert, T.; et al. The genomic diversification of the whole Acinetobacter genus: Origins, mechanisms, and consequences. Genome Biol. Evol. 2014, 10, 2866–2882. [Google Scholar] [CrossRef] [PubMed]
- Bearson, B.L.; Brunelle, B.W. Fluoroquinolone induction of phage-mediated gene transfer in multidrug-resistant Salmonella. Int. J. Antimicrob. Agents 2015, 46, 201–204. [Google Scholar] [CrossRef] [PubMed]
- Mašlaňová, I.; Stříbná, S.; Doškař, J.; Pantůček, R. Efficient plasmid transduction to Staphylococcus aureus strains insensitive to the lytic action of transducing phage. FEMS Microbiol. Lett. 2016, 363, fnw211. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Novick, R.P. Phage-mediated intergeneric transfer of toxin genes. Science 2009, 5910, 139–141. [Google Scholar] [CrossRef] [PubMed]
- Zeman, M.; Mašlaňová, I.; Indráková, A.; Šiborová, M.; Mikulášek, K.; Bendíčková, K.; Plevka, P.; Vrbovská, V.; Zdráhal, Z.; Doškař, J.; et al. Staphylococcus sciuri bacteriophages double-convert for staphylokinase and phospholipase, mediate interspecies plasmid transduction, and package mecA gene. Sci. Rep. 2017, 13, 46319. [Google Scholar] [CrossRef] [PubMed]
- Moon, B.Y.; Park, J.Y.; Robinson, D.A.; Thomas, J.C.; Park, Y.H.; Thornton, J.A.; Seo, K.S. Mobilization of Genomic Islands of Staphylococcus aureus by Temperate Bacteriophage. PLoS ONE 2016, 11, e0151409. [Google Scholar] [CrossRef] [PubMed]
- Matilla, M.A.; Salmond, G.P. Bacteriophage ϕMAM1, a viunalikevirus, is a broad-host-range, high-efficiency generalized transducer that infects environmental and clinical isolates of the enterobacterial genera Serratia and Kluyvera. Appl. Environ. Microbiol. 2014, 80, 6446–6457. [Google Scholar] [CrossRef] [PubMed]
- Varga, M.; Kuntová, L.; Pantůček, R.; Mašlaňová, I.; Růžičková, V.; Doškař, J. Efficient transfer of antibiotic resistance plasmids by transduction within methicillin-resistant Staphylococcus aureus USA300 clone. FEMS Microbiol. Lett. 2012, 332, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Valero-Rello, A.; López-Sanz, M.; Quevedo-Olmos, A.; Sorokin, A.; Ayora, S. Molecular Mechanisms That Contribute to Horizontal Transfer of Plasmids by the Bacteriophage SPP1. Front. Microbiol. 2017, 22, 1816. [Google Scholar] [CrossRef] [PubMed]
- Krahn, T.; Wibberg, D.; Maus, I.; Winkler, A.; Bontron, S.; Sczyrba, A.; Nordmann, P.; Pühler, A.; Poirel, L.; Schlüter, A. Intraspecies Transfer of the Chromosomal Acinetobacter baumannii blaNDM-1 Carbapenemase Gene. Antimicrob. Agents Chemother. 2016, 60, 3032–3040. [Google Scholar] [CrossRef] [PubMed]
- Touchon, M.; Bernheim, A.; Rocha, E.P. Genetic and life-history traits associated with the distribution of prophages in bacteria. ISME J. 2016, 11, 2744–2754. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, A.J.; Witney, A.A.; Lindsay, J.A. Staphylococcus aureus temperate bacteriophage: Carriage and horizontal gene transfer is lineage associated. Front. Cell. Infect. Microbiol. 2012, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Fogg, P.C.; Saunders, J.R.; McCarthy, A.J.; Allison, H.E. Cumulative effect of prophage burden on Shiga toxin production in Escherichia coli. Microbiology 2012, 158, 488–497. [Google Scholar] [CrossRef] [PubMed]
- Boyd, E.F.; Brüssow, H. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 2002, 11, 521–529. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, X.; Zhang, X.; Fu, J.; Wang, Z.; Chen, T.; Zhao, X. Characterization of genome-reduced Bacillus subtilis strains and their application for the production of guanosine and thymidine. Microb. Cell Fact. 2016, 15, 94. [Google Scholar] [CrossRef] [PubMed]
- Solheim, M.; Brekke, M.C.; Snipen, L.G.; Willems, R.J.; Nes, I.F.; Brede, DA. Comparative genomic analysis reveals significant enrichment of mobile genetic elements and genes encoding surface structure-proteins in hospital-associated clonal complex 2 Enterococcus faecalis. BMC Microbiol. 2011, 11, 3. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, I.T.; Banerjei, L.; Myers, G.S.; Nelson, K.E.; Seshadri, R.; Read, T.D.; Fouts, D.E.; Eisen, J.A.; Gill, S.R.; Heidelberg, J.F.; et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 2003, 5615, 2071–2074. [Google Scholar] [CrossRef] [PubMed]
- McBride, S.M.; Fischetti, V.A.; Leblanc, D.J.; Moellering, R.C., Jr.; Gilmore, M.S. Genetic diversity among Enterococcus faecalis. PLoS ONE 2007, 2, e582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matos, R.C.; Lapaque, N.; Rigottier-Gois, L.; Debarbieux, L.; Meylheuc, T.; Gonzalez-Zorn, B.; Repoila, F.; Lopes Mde, F.; Serror, P. Enterococcus faecalis prophage dynamics and contributions to pathogenic traits. PLoS Genet. 2013, 9, e1003539. [Google Scholar] [CrossRef] [PubMed]
- Goerke, C.; Pantucek, R.; Holtfreter, S.; Schulte, B.; Zink, M.; Grumann, D.; Bröker, B.M.; Doskar, J.; Wolz, C. Diversity of prophages in dominant Staphylococcus aureus clonal lineages. J. Bacteriol. 2009, 191, 3462–3468. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, F.; Bouzari, M.; Katouli, M.; Pourshafie, M.R. Prophage and antibiotic resistance profiles of methicillin-resistant Staphylococcus aureus strains in Iran. Arch. Virol. 2012, 157, 1807–1811. [Google Scholar] [CrossRef] [PubMed]
- Satta, G.; Pruzzo, C.; Debbia, E.; Fontana, R. Close association between shape alteration and loss of immunity to superinfection in a wild-type Klebsiella pneumoniae stable lysogen which can be both immune and nonimmune to superinfection. J. Virol. 1978, 28, 772–785. [Google Scholar] [PubMed]
- Kwon, T.; Jung, Y.H.; Lee, S.; Yun, M.R.; Kim, W.; Kim, D.W. Comparative genomic analysis of Klebsiella pneumoniae subsp. pneumoniae KP617 and PittNDM01, NUHL24835, and ATCC BAA-2146 reveals unique evolutionary history of this strain. Gut Pathog. 2016, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Wang, G.; Sebra, R.; Zhuge, J.; Yin, C.; Aguero-Rosenfeld, M.E.; Schuetz, A.N.; Dimitrova, N.; Fallon, J.T. Emergence and Evolution of Multidrug-Resistant Klebsiella pneumoniae with both bla(KPC) and bla(CTX-M) Integrated in the Chromosome. Antimicrob. Agents Chemother. 2017, 61, e00076-17. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xie, Y.; Li, G.; Liu, J.; Li, X.; Tian, L.; Sun, J.; Ou, H.Y.; Qu, H. Whole-Genome-Sequencing characterization of bloodstream infection-causing hypervirulent Klebsiella pneumoniae of capsular serotype K2 and ST374. Virulence 2018, 1, 510–521. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chavda, K.D.; DeLeo, F.R.; Bryant, K.A.; Jacobs, M.R.; Bonomo, R.A.; Kreiswirth, B.N. Genome Sequence of a Klebsiella pneumoniae Sequence Type 258 Isolate with Prophage-Encoded K. pneumoniae Carbapenemase. Genome Announc. 2015, 3, e00659-15. [Google Scholar] [CrossRef] [PubMed]
- Bi, D.; Jiang, X.; Sheng, Z.K.; Ngmenterebo, D.; Tai, C.; Wang, M.; Deng, Z.; Rajakumar, K.; Ou, H.Y. Mapping the resistance-associated mobilome of a carbapenem-resistant Klebsiella pneumoniae strain reveals insights into factors shaping these regions and facilitates generation of a ‘resistance-disarmed’ model organism. J. Antimicrob. Chemother. 2015, 10, 2770–2774. [Google Scholar] [CrossRef] [PubMed]
- Zautner, A.E.; Bunk, B.; Pfeifer, Y.; Spröer, C.; Reichard, U.; Eiffert, H.; Scheithauer, S.; Groß, U.; Overmann, J.; Bohne, W. Monitoring microevolution of OXA-48-producing Klebsiella pneumoniae ST147 in a hospital setting by SMRT sequencing. J. Antimicrob. Chemother. 2017, 72, 2737–2744. [Google Scholar] [CrossRef] [PubMed]
- Di Nocera, P.P.; Rocco, F.; Giannouli, M.; Triassi, M.; Zarrilli, R. Genome organization of epidemic Acinetobacter baumannii strains. BMC Microbiol. 2011, 11, 224. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, M.; Kurokawa, K.; Hayashi, T. Diversification of Escherichia coli genomes: Are bacteriophages the major contributors? Trends Microbiol. 2001, 10, 481–485. [Google Scholar] [CrossRef]
- Fouts, D.E. Phage_Finder: Automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res. 2006, 34, 5839–5851. [Google Scholar] [CrossRef] [PubMed]
- Ptashne, M. Genetic Switch: Phage Lambda and Higher Organisms, 2nd ed.; Blackwell: Cambridge, MA, USA, 1992. [Google Scholar]
- Cavalcanti, S.M.; Siqueira, J.P., Jr. Cure of prophage in Staphylococcus aureus by furocoumarin photoadditions. Microbios 1995, 327, 85–91. [Google Scholar]
- Duval-Iflah, Y. Lysogenic conversion of the lipase gene in Staphylococcus pyogenes group III strains. Can. J. Microbiol. 1972, 18, 1491–1497. [Google Scholar] [CrossRef] [PubMed]
- Gasson, M.J.; Davies, F.L. Prophage-cured derivatives of Streptococcus lactis and Streptococcus cremoris. Appl. Environ. Microbiol. 1980, 40, 964–966. [Google Scholar] [PubMed]
- Waldor, M.K.; Friedma, D.I. Phage regulatory circuits and virulence gene expression. Curr. Opin. Microbiol. 2005, 8, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Raya, R.R.; H’bert, E.M. Isolation of phage via induction of lysogens. Methods Mol. Biol. 2009, 501, 23–32. [Google Scholar] [PubMed]
- Selva, L.; Viana, D.; Regev-Yochay, G.; Trzcinski, K.; Corpa, J.M.; Lasa, I.; Novick, R.P.; Penadés, J.R. Killing niche competitors by remote-control bacteriophage induction. Proc. Natl. Acad. Sci. USA 2009, 106, 1234–1238. [Google Scholar] [CrossRef] [PubMed]
- Banks, D.J.; Lei, B.; Musser, J.M. Prophage induction and expression of prophage-encoded virulence factors in group A Streptococcus serotype M3 strain MGAS315. Infect. Immun. 2003, 71, 7079–7086. [Google Scholar] [CrossRef] [PubMed]
- Bertani, G. Studies on lysogenesis. III. Superinfection of lysogenic Shigella dysenteriae with temperate mutants of the carried phage. J. Bacteriol. 1954, 67, 696–707. [Google Scholar] [PubMed]
- Madera, C.; García, P.; Rodríguez, A.; Suárez, J.E.; Martínez, B. Prophage induction in Lactococcus lactis by the bacteriocin Lactococcin 972. Int. J. Food Microbiol. 2009, 129, 99–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Affolter, M.; Parent-Vaugeois, C.; Anderson, A. Curing and induction of the Fels 1 and Fels 2 prophages in the Ames mutagen tester strains of Salmonella typhimurium. Mutat. Res. 1983, 110, 243–262. [Google Scholar] [CrossRef]
- Menouni, R.; Champ, S.; Espinosa, L.; Boudvillain, M.; Ansaldi, M. Transcription termination controls prophage maintenance in Escherichia coli genomes. Proc. Natl. Acad. Sci. USA 2013, 110, 14414–14419. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.K.; Looft, T.; Bayles, D.O.; Humphrey, S.; Levine, U.Y.; Alt, D.; Stanton, T.B. Antibiotics in feed induce prophages in swine fecal microbiomes. mBio 2011, 6, e00260-11. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.; Roy, K.; Williamson, K.E.; Srinivasiah, S.; Wommack, K.E.; Radosevich, M. Acyl-homoserine lactones can induce virus production in lysogenic bacteria: An alternative paradigm for prophage induction. Appl. Environ. Microbiol. 2009, 75, 7142–7152. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.T.; Xavier, K.B.; Campagna, S.R.; Taga, M.E.; Semmelhack, M.F.; Bassler, B.L.; Hughson, F.M. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol. Cell 2004, 15, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Rossmann, F.S.; Racek, T.; Wobser, D.; Puchalka, J.; Rabener, E.M.; Reiger, M.; Hendrickx, A.P.; Diederich, A.K.; Jung, K.; Klein, C.; et al. Phage-mediated dispersal of biofilm and distribution of bacterial virulence genes is induced by quorum sensing. PLoS Pathog. 2015, 11, e1004653. [Google Scholar] [CrossRef] [PubMed]
- Lwoff, A. Lysogeny. Bacteriol. Rev. 1953, 17, 269–337. [Google Scholar] [PubMed]
- Birdsell, D.C.; Hathaway, G.M.; Rutberg, L. Characterization of Temperate Bacillus Bacteriophage phi105. J. Virol. 1969, 4, 264–270. [Google Scholar] [PubMed]
- Garro, A.J.; Law, M.F. Relationship between lysogeny, spontaneous induction, and transformation efficiencies in Bacillus subtilis. J. Bacteriol. 1974, 120, 1256–1259. [Google Scholar] [PubMed]
- Livny, J.; Friedman, D.I. Characterizing spontaneous induction of Stx encoding phages using a selectable reporter system. Mol. Microbiol. 2004, 51, 1691–1704. [Google Scholar] [CrossRef] [PubMed]
- Bullwinkle, T.J.; Koudelka, G.B. The lysis-lysogeny decision of bacteriophage 933W: A 933W repressor-mediated long-distance loop has no role in regulating 933W P(RM) activity. J. Bacteriol. 2011, 193, 3313–3323. [Google Scholar] [CrossRef] [PubMed]
- Carrolo, M.; Frias, M.J.; Pinto, F.R.; Melo-Cristino, J.; Ramirez, M. Prophage spontaneous activation promotes DNA release enhancing biofilm formation in Streptococcus pneumoniae. PLoS ONE 2010, 5, e15678. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Kim, Y.; Ma, Q.; Hong, S.H.; Pokusaeva, K.; Sturino, J.M.; Wood, T.K. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 2010, 1, 147. [Google Scholar] [CrossRef] [PubMed]
- Nanda, A.M.; Heyer, A.; Krämer, C.; Grünberger, A.; Kohlheyer, D.; Frunzke, J. Analysis of SOS-induced spontaneous prophage induction in Corynebacterium glutamicum at the single-cell level. J. Bacteriol. 2014, 196, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Rippon, J.E. The classification of bacteriophages lysing staphylococci. J. Hyg. 1956, 54, 213–226. [Google Scholar] [CrossRef] [PubMed]
- Allué-Guardia, A.; García-Aljaro, C.; Muniesa, M. Bacteriophage-encoding cytolethal distending toxin type V gene induced from nonclinical Escherichia coli isolates. Infect. Immun. 2011, 79, 3262–3272. [Google Scholar] [CrossRef]
- Zhou, Y.; Liang, Y.; Lynch, K.H.; Dennis, J.J.; Wishart, D.S. PHAST: A fast phage search tool. Nucleic Acids Res. 2011, 39, W347–W352. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; Marcu, A.; Liang, Y.; Wishart, D.S. PHAST, PHASTER and PHASTEST: Tools for finding prophage in bacterial genomes. Brief Bioinform. 2017. [Google Scholar] [CrossRef] [PubMed]
- Lima-Mendez, G.; Van Helden, J.; Toussaint, A.; Leplae, R. Prophinder: A computational tool for prophage prediction in prokaryotic genomes. Bioinformatics 2008, 24, 863–865. [Google Scholar] [CrossRef] [PubMed]
- Bose, M.; Barber, R.D. Prophage Finder: A prophage loci prediction tool for prokaryotic genome sequences. In Silico Biol. 2006, 6, 223–227. [Google Scholar] [PubMed]
- Akhter, S.; Aziz, R.K.; Edwards, R.A. PhiSpy: A novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res. 2012, 40, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Enault, F.; Hurwitz, B.L.; Sullivan, M.B. VirSorter: Mining viral signal from microbial genomic data. PeerJ 2015, 3, e985. [Google Scholar] [CrossRef] [PubMed]
- Pantůcek, R.; Doskar, J.; Růzicková, V.; Kaspárek, P.; Orácová, E.; Kvardová, V.; Rosypal, S. Identification of bacteriophage types and their carriage in Staphylococcus aureus. Arch. Virol. 2004, 149, 1689–1703. [Google Scholar] [CrossRef] [PubMed]
- Kahánková, J.; Pantůček, R.; Goerke, C.; Růžičková, V.; Holochová, P.; Doškař, J. Multilocus PCR typing strategy for differentiation of Staphylococcus aureus siphoviruses reflecting their modular genome structure. Environ. Microbiol. 2010, 12, 2527–2538. [Google Scholar] [CrossRef] [PubMed]
- Ross, I.L.; Heuzenroeder, M.W. Discrimination within phenotypically closely related definitive types of Salmonella enterica serovar typhimurium by the multiple amplification of phage locus typing technique. J. Clin. Microbiol. 2005, 43, 1604–1611. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.Y.; Iandolo, J.J. Mechanism of bacteriophage conversion of lipase activity in Staphylococcus aureus. J. Bacteriol. 1985, 164, 288–293. [Google Scholar] [PubMed]
- Loeffler, J.M.; Fischetti, V.A. Lysogeny of Streptococcus pneumoniae with MM1 phage: Improved adherence and other phenotypic changes. Infect. Immun. 2006, 74, 4486–4495. [Google Scholar] [CrossRef] [PubMed]
- Leffers, G.G., Jr.; Gottesman, S. Lambda Xis degradation in vivo by Lon and FtsH. J. Bacteriol. 1998, 180, 1573–1577. [Google Scholar] [PubMed]
- Panis, G.; Méjean, V.; Ansaldi, M. Control and regulation of KplE1 prophage site-specific recombination: A new recombination module analyzed. J. Biol. Chem. 2007, 282, 21798–21809. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Bossi, N.; Coissac, E.; Netter, P.; Bossi, L. Unsuspected prophage-like elements in Salmonella typhimurium. Mol. Microbiol. 1997, 25, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Bae, T.; Baba, T.; Hiramatsu, K.; Schneewind, O. Prophages of Staphylococcus aureus Newman and their contribution to virulence. Mol. Microbiol. 2006, 62, 1035–1047. [Google Scholar] [CrossRef] [PubMed]
- Koskella, B.; Meaden, S. Understanding bacteriophage specificity in natural microbial communities. Viruses 2013, 5, 806–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyman, P.; Abedon, S.T. Bacteriophage host range and bacterial resistance. Adv. Appl. Microbiol. 2010, 70, 217–248. [Google Scholar] [CrossRef] [PubMed]
- Rakhuba, D.V.; Kolomiets, E.I.; Dey, E.S.; Novik, G.I. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol. J. Microbiol. 2010, 59, 145–155. [Google Scholar] [PubMed]
- Xia, G.; Maier, L.; Sanchez-Carballo, P.; Li, M.; Otto, M.; Holst, O.; Peschel, A. Glycosylation of wall teichoic acid in Staphylococcus aureus by TarM. J. Biol. Chem. 2010, 285, 13405–13415. [Google Scholar] [CrossRef] [PubMed]
- Winstel, V.; Xia, G.; Peschel, A. Pathways and roles of wall teichoic acid glycosylation in Staphylococcus aureus. Int. J. Med. Microbiol. 2014, 304, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Dutta, V.; Elhanafi, D.; Lee, S.; Osborne, J.A.; Kathariou, S. A novel restriction-modification system is responsible for temperature-dependent phage resistance in Listeria monocytogenes ECII. Appl. Environ. Microbiol. 2012, 78, 1995–2004. [Google Scholar] [CrossRef] [PubMed]
- Dowah, A.S.A.; Clokie, M.R.J. Review of the nature, diversity and structure of bacteriophage receptor binding proteins that target Gram-positive bacteria. Biophys. Rev. 2018. [Google Scholar] [CrossRef] [PubMed]
- Bertozzi, S.J.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016, 363, fnw002. [Google Scholar] [CrossRef] [PubMed]
- Ainsworth, S.; Sadovskaya, I.; Vinogradov, E.; Courtin, P.; Guerardel, Y.; Mahony, J.; Grard, T.; Cambillau, C.; Chapot-Chartier, M.P.; van Sinderen, D. Differences in lactococcal cell wall polysaccharide structure are major determining factors in bacteriophage sensitivity. mBio 2014, 6, e00880-14. [Google Scholar] [CrossRef] [PubMed]
- Ardissone, S.; Fumeaux, C.; Bergé, M.; Beaussart, A.; Théraulaz, L.; Radhakrishnan, S.K.; Dufrêne, YF.; Viollier, P.H. Cell cycle constraints on capsulation and bacteriophage susceptibility. Elife 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Dy, R.L.; Richter, C.; Salmond, G.P.; Fineran, P.C. Remarkable Mechanisms in Microbes to Resist Phage Infections. Annu. Rev. Virol. 2014, 1, 307–331. [Google Scholar] [CrossRef] [PubMed]
- Seed, K.D. Battling Phages: How Bacteria Defend against Viral Attack. PloS Pathog. 2015, 11, e1004847. [Google Scholar] [CrossRef] [PubMed]
- Zschach, H.; Larsen, M.V.; Hasman, H.; Westh, H.; Nielsen, M.; Międzybrodzki, R.; Jończyk-Matysiak, E.; Weber-Dąbrowska, B.; Górski, A. Use of a Regression Model to Study Host-Genomic Determinants of Phage Susceptibility in MRSA. Antibiotics 2018, 7, E9. [Google Scholar] [CrossRef] [PubMed]
- Shabbir, M.A.; Hao, H.; Shabbir, M.Z.; Wu, Q.; Sattar, A.; Yuan, Z. Bacteria vs. Bacteriophages: Parallel Evolution of Immune Arsenals. Front. Microbiol. 2016, 7, 1292. [Google Scholar] [CrossRef] [PubMed]
- Goldfarb, T.; Sberro, H.; Weinstock, E.; Cohen, O.; Doron, S.; Charpak-Amikam, Y.; Afik, S.; Ofir, G.; Sorek, R. BREX is a novel phage resistance system widespread in microbial genomes. EMBO J. 2015, 34, 169–183. [Google Scholar] [CrossRef] [PubMed]
- Chopin, M.C.; Chopin, A.; Bidnenko, E. Phage abortive infection in lactococci: Variations on a theme. Curr. Opin. Microbiol. 2005, 8, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gerlach, D.; Du, X.; Larsen, J.; Stegger, M.; Kühner, P.; Peschel, A.; Xia, G.; Winstel, V. An accessory wall teichoic acid glycosyltransferase protects Staphylococcus aureus from the lytic activity of Podoviridae. Sci. Rep. 2015, 5, 17219. [Google Scholar] [CrossRef] [PubMed]
- Tzipilevich, E.; Habusha, M.; Ben-Yehuda, S. Acquisition of Phage Sensitivity by Bacteria through Exchange of Phage Receptors. Cell 2017, 168, 186.e2–199.e2. [Google Scholar] [CrossRef] [PubMed]
- Deng, K.; Fang, W.; Zheng, B.; Miao, S.; Huo, G. Phenotypic, fermentation characterization, and resistance mechanism analysis of bacteriophage-resistant mutants of Lactobacillus delbrueckiissp. bulgaricus isolated from traditional Chinese dairy products. J. Dairy Sci. 2018, 101, 1901–1914. [Google Scholar] [CrossRef] [PubMed]
- Zago, M.; Orrù, L.; Rossetti, L.; Lamontanara, A.; Fornasari, M.E.; Bonvini, B.; Meucci, A.; Carminati, D.; Cattivelli, L.; Giraffa, G. Survey on the phage resistance mechanisms displayed by a dairy Lactobacillus helveticus strain. Food Microbiol. 2017, 66, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Suárez, V.B.; Maciel, N.; Guglielmotti, D.; Zago, M.; Giraffa, G.; Reinheimer, J. Phage-resistance linked to cell heterogeneity in the commercial strain Lactobacillus delbrueckii subsp. lactis Ab1. Int. J. Food Microbiol. 2008, 128, 401–405. [Google Scholar] [CrossRef]
- Koczula, A.; Willenborg, J.; Bertram, R.; Takamatsu, D.; Valentin-Weigand, P.; Goethe, R. Establishment of a Cre recombinase based mutagenesis protocol for markerless gene deletion in Streptococcus suis. J. Microbiol. Methods 2014, 107, 80–83. [Google Scholar] [CrossRef] [PubMed]
- Zaleski, P.; Wojciechowski, M.; Piekarowicz, A. The role of Dam methylation in phase variation of Haemophilus influenzae genes involved in defence against phage infection. Microbiology 2005, 151, 3361–3369. [Google Scholar] [CrossRef] [PubMed]
- Styriak, I.; Pristas, P.; Javorský, P. Lack of surface receptors not restriction-modification system determines F4 phage resistance in Streptococcus bovis II/1. Folia Microbiol. 1998, 43, 35–38. [Google Scholar] [CrossRef]
- Sanders, M.E. Phage resistance in lactic acid bacteria. Biochimie 1988, 70, 411–422. [Google Scholar] [CrossRef]
- Faruque, S.M.; Bin Naser, I.; Fujihara, K.; Diraphat, P.; Chowdhury, N.; Kamruzzaman, M.; Qadri, F.; Yamasaki, S.; Ghosh, A.N.; Mekalanos, J.J. Genomic sequence and receptor for the Vibrio cholerae phage KSF-1phi: Evolutionary divergence among filamentous vibriophages mediating lateral gene transfer. J. Bacteriol. 2005, 187, 4095–4103. [Google Scholar] [CrossRef] [PubMed]
- Smarda, J.; Doroszkiewicz, W.; Lachowicz, T.M. Sensitivity of Shigella flexneri and Escherichia coli bacteria to bacteriophages and to colicins, lost or established by the acquisition of R plasmids. Acta Microbiol. Pol. 1990, 39, 23–35. [Google Scholar] [PubMed]
- Ram, G.; Chen, J.; Ross, H.F.; Novick, R.P. Precisely modulated pathogenicity island interference with late phage gene transcription. Proc. Natl. Acad. Sci. USA 2014, 111, 14536–14541. [Google Scholar] [CrossRef] [PubMed]
- Hofer, B.; Ruge, M.; Dreiseikelmann, B. The superinfection exclusion gene (sieA) of bacteriophage P22: Identification and overexpression of the gene and localization of the gene product. J. Bacteriol. 1995, 177, 3080–3086. [Google Scholar] [CrossRef] [PubMed]
- Dempsey, R.M.; Carroll, D.; Kong, H.; Higgins, L.; Keane, C.T.; Coleman, D.C. Sau42I, a BcgI-like restriction-modification system encoded by the Staphylococcus aureus quadruple-converting phage Phi42. Microbiology 2005, 151, 1301–1311. [Google Scholar] [CrossRef] [PubMed]
- Coleman, D.C.; Sullivan, D.J.; Russell, R.J.; Arbuthnott, J.P.; Carey, B.F.; Pomeroy, H.M. Staphylococcus aureus bacteriophages mediating the simultaneous lysogenic conversion of beta-lysin, staphylokinase and enterotoxin A: Molecular mechanism of triple conversion. J. Gen. Microbiol. 1989, 135, 1679–1697. [Google Scholar] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Snir, S.; Koonin, E.V. Defense islands in bacterial and archaeal genomes and prediction of novel defense systems. J. Bacteriol. 2011, 193, 6039–6056. [Google Scholar] [CrossRef] [PubMed]
- O’Driscoll, J.; Glynn, F.; Cahalane, O.; O’Connell-Motherway, M.; Fitzgerald, G.F.; Van Sinderen, D. Lactococcal Plasmid pNP40 Encodes a Novel, Temperature-Sensitive Restriction-Modification System. Appl. Environ. Microbiol. 2004, 70, 5546–5556. [Google Scholar] [CrossRef] [PubMed]
- Trotter, M.; Ross, R.P.; Fitzgerald, G.F.; Coffey, A. Lactococcus lactis DPC5598, a plasmid-free derivative of a commercial starter, provides a valuable alternative host for culture improvement studies. J. Appl. Microbiol. 2002, 93, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Boucher, I.; Emond, E.; Parrot, M.; Moineau, S. DNA sequence analysis of three Lactococcus lactis plasmids encoding phage resistance mechanisms. J. Dairy Sci. 2001, 84, 1610–1620. [Google Scholar] [CrossRef]
- Burrus, V.; Bontemps, C.; Decaris, B.; Guédon, G. Characterization of a novel type II restriction-modification system, Sth368I, encoded by the integrative element ICESt1 of Streptococcus thermophilus CNRZ368. Appl. Environ. Microbiol. 2001, 67, 1522–1528. [Google Scholar] [CrossRef] [PubMed]
- Forde, A.; Daly, C.; Fitzgerald, G.F. Identification of four phage resistance plasmids from Lactococcus lactis subsp. cremoris HO2. Appl. Environ. Microbiol. 1999, 65, 1540–1547. [Google Scholar] [PubMed]
- Mohammed, M.; Cormican, M. Whole genome sequencing provides possible explanations for the difference in phage susceptibility among two Salmonella Typhimurium phage types (DT8 and DT30) associated with a single foodborne outbreak. BMC 2015, 8, 728. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.M.; Harvey, M.L.; Liu, C.Q.; Dunn, N.W. A novel plasmid-encoded phage abortive infection system from Lactococcus lactis biovar. diacetylactis. FEMS Microbiol Lett. 1997, 146, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Millen, A.M.; Horvath, P.; Boyaval, P.; Romero, D.A. Mobile CRISPR/Cas-mediated bacteriophage resistance in Lactococcus lactis. PLoS ONE 2012, 12, e51663. [Google Scholar] [CrossRef] [PubMed]
- Tock, M.R.; Dryden, D.T. The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 2005, 8, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Ofir, G.; Sorek, R. Contemporary Phage Biology: From Classic Models to New Insights. Cell 2018, 172, 1260–1270. [Google Scholar] [CrossRef] [PubMed]
- Samson, J.E.; Magadán, A.H.; Sabri, M.; Moineau, S. Revenge of the phages: Defeating bacterial defences. Nat. Rev. Microbiol. 2013, 11, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Suárez, V.; Zago, M.; Giraffa, G.; Reinheimer, J.; Quiberoni, A. Evidence for the presence of restriction/modification systems in Lactobacillus delbrueckii. J. Dairy Res. 2009, 76, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Akatov, A.K.; Zueva, V.S.; Dmitrenko, O.A. A new approach to establishing the set of phages for typing methicillin-resistant Staphylococcus aureus. J. Chemother. 1991, 5, 275–278. [Google Scholar] [CrossRef]
- Waldron, D.E.; Lindsay, J.A. Sau1: A novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. aureus isolates of different lineages. J. Bacteriol. 2006, 188, 5578–5585. [Google Scholar] [CrossRef] [PubMed]
- Corvaglia, A.R.; François, P.; Hernandez, D.; Perron, K.; Linder, P.; Schrenzel, J. A type III-like restriction endonuclease functions as a major barrier to horizontal gene transfer in clinical Staphylococcus aureus strains. Proc. Natl. Acad. Sci. USA 2010, 107, 11954–11958. [Google Scholar] [CrossRef] [PubMed]
- Cady, K.C.; Bondy-Denomy, J.; Heussler, G.E.; Davidson, A.R.; O’Toole, G.A. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J. Bacteriol. 2012, 194, 5728–5738. [Google Scholar] [CrossRef] [PubMed]
- Fineran, P.C.; Blower, T.R.; Foulds, I.J.; Humphreys, D.P.; Lilley, K.S.; Salmond, G.P. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl. Acad. Sci. USA 2009, 106, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Haaber, J.; Moineau, S.; Fortier, L.C.; Hammer, K. AbiV, a novel antiphage abortive infection mechanism on the chromosome of Lactococcus lactis subsp. cremoris MG1363. Appl. Environ. Microbiol. 2008, 74, 6528–6537. [Google Scholar] [CrossRef] [PubMed]
- Durmaz, E.; Klaenhammer, T.R. Genetic analysis of chromosomal regions of Lactococcus lactis acquired by recombinant lytic phages. Appl. Environ. Microbiol. 2000, 66, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Adams, B.L. The Next Generation of Synthetic Biology Chassis: Moving Synthetic Biology from the Laboratory to the Field. ACS Synth. Biol. 2016, 12, 1328–1330. [Google Scholar] [CrossRef] [PubMed]
- Szathmary, E. Life—In search of the simplest cell. Nature 2005, 433, 469–470. [Google Scholar] [CrossRef] [PubMed]
- Umenhoffer, K.; Draskovits, G.; Nyerges, Á.; Karcagi, I.; Bogos, B.; Tímár, E.; Csörgő, B.; Herczeg, R.; Nagy, I.; Fehér, T.; et al. Genome-Wide Abolishment of Mobile Genetic Elements Using Genome Shuffling and CRISPR/Cas-Assisted MAGE Allows the Efficient Stabilization of a Bacterial Chassis. ACS Synth. Biol. 2017, 8, 1471–1483. [Google Scholar] [CrossRef] [PubMed]
- Lauritsen, I.; Porse, A.; Sommer, M.O.A.; Nørholm, M.H.H. A versatile one-step CRISPR-Cas9 based approach to plasmid-curing. Microb. Cell Fact. 2017, 16, 135. [Google Scholar] [CrossRef] [PubMed]
- Ellis, H.M.; Yu, D.; DiTizio, T.; Court, D.L. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc. Natl. Acad. Sci. USA 2001, 98, 6742–6746. [Google Scholar] [CrossRef] [PubMed]
- Martínez-García, E.; de Lorenzo, V. Molecular tools and emerging strategies for deep genetic/genomic refactoring of Pseudomonas. Curr. Opin. Biotechnol. 2017, 47, 120–132. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.H.; Isaacs, F.J.; Carr, P.A.; Sun, Z.Z.; Xu, G.; Forest, C.R.; Church, G.M. Programming cells by multiplex genome engineering and accelerated evolution. Nature 2009, 7257, 894–898. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Xing, X.H.; Zhang, C. Targeted mutagenesis: A sniper-like diversity generator in microbial engineering. Synth. Syst. Biotechnol. 2017, 2, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Ellis, H.M.; Lee, E.C.; Jenkins, N.A.; Copeland, N.G.; Court, D.L. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 2000, 97, 5978–5983. [Google Scholar] [CrossRef] [PubMed]
- Zerbini, F.; Zanella, I.; Fraccascia, D.; König, E.; Irene, C.; Frattini, L.F.; Tomasi, M.; Fantappiè, L.; Ganfini, L.; Caproni, E.; et al. Large scale validation of an efficient CRISPR/Cas-based multi gene editing protocol in Escherichia coli. Microb. Cell Fact. 2017, 16, 68. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Jiang, Y.; Shao, L.; Yang, P.; Sun, B.; Yang, S.; Chen, D. CRISPR/Cas9-based efficient genome editing in Staphylococcus aureus. Acta Biochim. Biophys. Sin. 2017, 49, 764–770. [Google Scholar] [CrossRef] [PubMed]
- Ricaurte, D.E.; Martínez-García, E.; Nyerges, Á.; Pál, C.; de Lorenzo, V.; Aparicio, T. A standardized workflow for surveying recombinases expands bacterial genome-editing capabilities. Microb. Biotechnol. 2018, 11, 176–188. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, T.; Jensen, S.I.; Nielsen, A.T.; de Lorenzo, V.; Martinez-Garcia, E. The Ssr protein (T1E_1405) from Pseudomonas putida DOT-T1E enables oligonucleotide-based recombineering in platform strain P. putida EM42. Biotechnol. J. 2016, 11, 1309–1319. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ram, G.; Yoong, P.; Penadés, J.R.; Shopsin, B.; Novick, R.P. An rpsL-based allelic exchange vector for Staphylococcus aureus. Plasmid 2015, 79, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Prax, M.; Lee, C.Y.; Bertram, R. An update on the molecular genetics toolbox for staphylococci. Microbiology 2013, 159, 421–435. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tian, P. Contemplating 3-Hydroxypropionic Acid Biosynthesis in Klebsiella pneumoniae. Indian J. Microbiol. 2015, 55, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Penewit, K.; Holmes, E.A.; McLean, K.; Ren, M.; Waalkes, A.; Salipante, S.J. Efficient and Scalable Precision Genome Editing in Staphylococcus aureus through Conditional Recombineering and CRISPR/Cas9-Mediated Counterselection. mBio 2010, 9, e00067-18. [Google Scholar] [CrossRef] [PubMed]
- Tucker, A.T.; Nowicki, E.M.; Boll, J.M.; Knauf, G.A.; Burdis, N.C.; Trent, M.S.; Davies, B.W. Defining gene-phenotype relationships in Acinetobacter baumannii through one-step chromosomal gene inactivation. mBio 2014, 5, e01313-14. [Google Scholar] [CrossRef] [PubMed]
- Dalia, T.N.; Yoon, S.H.; Galli, E.; Barre, F.X.; Waters, C.M.; Dalia, A.B. Enhancing multiplex genome editing by natural transformation (MuGENT) via inactivation of ssDNA exonucleases. Nucleic Acids Res. 2017, 45, 7527–7537. [Google Scholar] [CrossRef] [PubMed]
- Prathapam, R.; Uehara, T. A temperature-sensitive replicon enables efficient gene inactivation in Pseudomonas aeruginosa. J. Microbiol. Methods 2018, 144, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Pósfai, G.; Plunkett, G., 3rd; Fehér, T.; Frisch, D.; Keil, G.M.; Umenhoffer, K.; Kolisnychenko, V.; Stahl, B.; Sharma, S.S.; de Arruda, M.; et al. Emergent properties of reduced-genome Escherichia coli. Science 2006, 5776, 1044–1046. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Garcia, E.; Nikel, P.I.; Aparicio, T.; de Lorenzo, V. Pseudomonas 2.0: Genetic upgrading of P. putida KT2440 as an enhanced host for heterologous gene expression. Microb. Cell Fact. 2014, 13, 159. [Google Scholar] [CrossRef] [PubMed]
- Martínez-García, E.; Jatsenko, T.; Kivisaar, M.; de Lorenzo, V. Freeing Pseudomonas putida KT2440 of its proviral load strengthens endurance to environmental stresses. Environ. Microbiol. 2015, 17, 76–90. [Google Scholar] [CrossRef] [PubMed]
- Baumgart, M.; Unthan, S.; Rückert, C.; Sivalingam, J.; Grünberger, A.; Kalinowski, J.; Bott, M.; Noack, S.; Frunzke, J. Construction of a prophage-free variant of Corynebacterium glutamicum ATCC 13032 for use as a platform strain for basic research and industrial biotechnology. Appl. Environ. Microbiol. 2013, 79, 6006–6015. [Google Scholar] [CrossRef] [PubMed]
- Łobocka, M.; Gozdek, A.; Gozdek, A.; Izak, D.; Zalewska, A.; Gawor, J.; Dąbrowski, K.; Gromadka, R.; Weber-Dąbrowska, B.; Górski, A. Enterococcus faecalis strains for the production of Bacteriophage Preparations. PCT Patent Application WO 2016/030872 A1, 16 March 2016. [Google Scholar]
- Balogh, B.; Jones, J.B.; Iriarte, F.B.; Momol, M.T. Phage therapy for plant disease control. Curr. Pharm. Biotechnol. 2010, 11, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Gill, J.J.; Hyman, P. Phage choice, isolation, and preparation for phage therapy. Curr. Pharm. Biotechnol. 2010, 11, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Hatfull, G.F. The secret lives of mycobacteriophages. Adv. Virus Res. 2012, 82, 179–288. [Google Scholar] [PubMed]
- Carlton, R.M.; Noordman, W.H.; Biswas, B.; de Meester, E.D.; Loessner, M.J. Bacteriophage P100 for control of Listeria monocytogenes in foods: Genome sequence, bioinformatic analyses, oral toxicity study, and application. Regul. Toxicol. Pharm. 2005, 43, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Lehman, S.M.; Kropinski, A.M.; Castle, A.J.; Svircev, A.M. Complete genome of the broad-host-range Erwinia amylovora phage phiEa21-4 and its relationship to Salmonella phage Félix O1. Appl. Environ. Microbiol. 2009, 75, 2139–2147. [Google Scholar] [CrossRef] [PubMed]
- Santos, S.B.; Fernandes, E.; Carvalho, C.M.; Sillankorva, S.; Krylov, V.N.; Pleteneva, E.A.; Shaburova, O.V.; Nicolau, A.; Ferreira, E.C.; Azeredo, J. Selection and characterization of a multivalent Salmonella phage and its production in a nonpathogenic Escherichia coli strain. Appl. Environ. Microbiol. 2010, 76, 7338–7342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Haddad, L.; Ben Abdallah, N.; Plante, P.L.; Dumaresq, J.; Katsarava, R.; Labrie, S.; Corbeil, J.; St-Gelais, D.; Moineau, S. Improving the safety of Staphylococcus aureus polyvalent phages by their production on a Staphylococcus xylosus strain. PLoS ONE 2014, 9, e102600. [Google Scholar] [CrossRef] [PubMed]
- González-Menéndez, E.; Arroyo-López, F.N.; Martínez, B.; García, P.; Garrido-Fernández, A.; Rodríguez, A. Optimizing Propagation of Staphylococcus aureus Infecting Bacteriophage vB_SauM-phiIPLA-RODI on Staphylococcus xylosus Using Response Surface Methodology. Viruses 2018, 10, E153. [Google Scholar] [CrossRef] [PubMed]
- Karli, A.; Sensoy, G.; Unal, N.; Yanik, K.; Cigdem, H.; Belet, N.; Sofuoglu, A. Ventriculoperitoneal shunt infection with Listeria innocua. Pediatr. Int. 2014, 56, 621–623. [Google Scholar] [CrossRef] [PubMed]
- Moreno, L.Z.; Paixão, R.; Gobbi, D.D.; Raimundo, D.C.; Ferreira, T.P.; Hofer, E.; Matte, M.H.; Moreno, A.M. Characterization of atypical Listeria innocua isolated from swine slaughterhouses and meat markets. Res. Microbiol. 2012, 163, 268–271. [Google Scholar] [CrossRef] [PubMed]
- Reyrat, J.M.; Kahn, D. Mycobacterium smegmatis: An absurd model for tuberculosis? Trends Microbiol. 2001, 10, 472–474. [Google Scholar] [CrossRef]
- Büyükcam, A.; Tuncer, Ö.; Gür, D.; Sancak, B.; Ceyhan, M.; Cengiz, A.B.; Kara, A. Clinical and microbiological characteristics of Pantoea agglomerans infection in children. J. Infect. Public Health 2018, 11, 304–309. [Google Scholar] [CrossRef] [PubMed]
- Dutkiewicz, J.; Mackiewicz, B.; Kinga Lemieszek, M.; Golec, M.; Milanowski, J. Pantoea agglomerans: A mysterious bacterium of evil and good. Part III. Deleterious effects: Infections of humans, animals and plants. Ann. Agric. Environ. Med. 2016, 23, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Arora, A.; Sathyabama, S.; Mubin, N.; Verma, S.; Mayilraj, S.; Agrewala, J.N. Genome sequencing, assembly, annotation and analysis of Staphylococcus xylosus strain DMB3-Bh1 reveals genes responsible for pathogenicity. Gut Pathog. 2016, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Thornton, V.B.; Davis, J.A.; St Clair, M.B.; Cole, M.N. Inoculation of Staphylococcus xylosus in SJL/J mice to determine pathogenicity. Contemp. Top. Lab. Anim. Sci. 2003, 42, 49–52. [Google Scholar] [PubMed]
- Almeida, R.A.; Oliver, S.P. Interaction of coagulase-negative Staphylococcus species with bovine mammary epithelial cells. Microb. Pathog. 2001, 31, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Clayton, E.M.; Daly, K.M.; Guinane, C.M.; Hill, C.; Cotter, P.D.; Ross, P.R. Atypical Listeria innocua strains possess an intact LIPI-3. BMC Microbiol. 2014, 14, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno, L.Z.; Paixão, R.; de Gobbi, D.D.; Raimundo, D.C.; Porfida Ferreira, T.S.; Micke Moreno, A.; Hofer, E.; dos Reis, C.M.F.; Matté, G.R.; Matté, M.H. Phenotypic and genotypic characterization of atypical Listeria monocytogenes and Listeria innocua isolated from swine slaughterhouses and meat markets. Biomed. Res. Int. 2014, 2014, 742032. [Google Scholar] [CrossRef] [PubMed]
- Coros, A.; DeConno, E.; Derbyshire, K.M. IS6110, a Mycobacterium tuberculosis complex-specific insertion sequence, is also present in the genome of Mycobacterium smegmatis, suggestive of lateral gene transfer among mycobacterial species. J. Bacteriol. 2008, 190, 3408–3410. [Google Scholar] [CrossRef] [PubMed]
- Derbyshire, K.M.; Gray, T.A. Distributive Conjugal Transfer: New Insights into Horizontal Gene Transfer and Genetic Exchange in Mycobacteria. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Naum, M.; Brown, E.W.; Mason-Gamer, R.J. Phylogenetic evidence for extensive horizontal gene transfer of type III secretion system genes among enterobacterial plant pathogens. Microbiology 2009, 155, 3187–3199. [Google Scholar] [CrossRef] [PubMed]
- Kirzinger, M.W.; Butz, C.J.; Stavrinides, J. Inheritance of Pantoea type III secretion systems through both vertical and horizontal transfer. Mol. Genet. Genom. 2015, 290, 2075–2088. [Google Scholar] [CrossRef] [PubMed]
- Tormo, M.A.; Knecht, E.; Götz, F.; Lasa, I.; Penadés, J.R. Bap-dependent biofilm formation by pathogenic species of Staphylococcus: Evidence of horizontal gene transfer? Microbiology 2005, 151, 2465–2475. [Google Scholar] [CrossRef] [PubMed]
- Näsvall, J.; Knöppel, A.; Andersson, D.I. Duplication-Insertion Recombineering: A fast and scar-free method for efficient transfer of multiple mutations in bacteria. Nucleic Acids Res. 2017, 45, e33. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.; Schäfer, R.; Kohlstedt, M.; Harder, B.J.; Borchert, N.S.; Stöveken, N.; Bremer, E.; Wittmann, C. Systems metabolic engineering of Corynebacterium glutamicum for production of the chemical chaperone ectoine. Microb. Cell Fact. 2013, 12, 110. [Google Scholar] [CrossRef] [PubMed]
- Aubert, D.F.; Hamad, M.A.; Valvano, M.A. A markerless deletion method for genetic manipulation of Burkholderia cenocepacia and other multidrug-resistant gram-negative bacteria. Methods Mol. Biol. 2014, 1197, 311–327. [Google Scholar] [CrossRef] [PubMed]
- Unthan, S.; Baumgart, M.; Radek, A.; Herbst, M.; Siebert, D.; Brühl, N.; Bartsch, A.; Bott, M.; Wiechert, W.; Marin, K.; et al. Chassis organism from Corynebacterium glutamicum—A top-down approach to identify and delete irrelevant gene clusters. Biotechnol. J. 2015, 10, 290–301. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Tian, K.M.; Niu, D.D.; Shen, W.; Shi, G.Y.; Singh, S.; Wang, Z.X. Improvement of D-lactate productivity in recombinant Escherichia coli by coupling production with growth. Biotechnol. Lett. 2012, 34, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liu, Y.; Shin, H.D.; Chen, R.R.; Wang, N.S.; Li, J.; Du, G.; Chen, J. Developing Bacillus spp. as a cell factory for production of microbial enzymes and industrially important biochemicals in the context of systems and synthetic biology. Appl. Microbiol. Biotechnol. 2013, 97, 6113–6127. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Gao, W.; Feng, J.; Zhang, C.; He, Y.; Cao, M.; Li, Q.; Sun, Y.; Yang, C.; Song, C.; et al. A markerless gene replacement method for B. amyloliquefaciens LL3 and its use in genome reduction and improvement of poly-γ-glutamic acid production. Appl. Microbiol. Biotechnol. 2014, 98, 8963–8973. [Google Scholar] [CrossRef] [PubMed]
- Leprince, A.; van Passel, M.W.J.; Dos Santos, V.A.P.M. Streamlining genomes: Toward the generation of simplified and stabilized microbial systems. Curr. Opin. Biotechnol. 2012, 23, 651–658. [Google Scholar] [CrossRef] [PubMed]
- Lieder, S.; Nikel, P.I.; de Lorenzo, V.; Takors, R. Genome reduction boosts heterologous gene expression in Pseudomonas putida. Microb. Cell Fact. 2015, 14, 23. [Google Scholar] [CrossRef] [PubMed]
- Sabri, S.; Steen, J.A.; Bongers, M.; Nielsen, L.K.; Vickers, C.E. Knock-in/Knock-out (KIKO) vectors for rapid integration of large DNA sequences, including whole metabolic pathways, onto the Escherichia coli chromosome at well-characterised loci. Microb. Cell Fact. 2013, 12, 60. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.H.; Schweizer, H.P. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 2005, 5, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Zou, Z.; Kreth, J.; Merritt, J. Recombineering in Streptococcus mutans Using Direct Repeat-Mediated Cloning-Independent Markerless Mutagenesis (DR-CIMM). Front. Cell. Infect. Microbiol. 2017, 7, 202. [Google Scholar] [CrossRef] [PubMed]
- Bauer, R.; Mauerer, S.; Grempels, A.; Spellerberg, B. The competence system of Streptococcus anginosus and its use for genetic engineering. Mol. Oral Microbiol. 2018, 33, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.Y.; Yan, H.Q.; Ren, G.X.; Zhao, J.P.; Guo, X.P.; Sun, Y.C. CRISPR-Cas12a-Assisted Recombineering in Bacteria. Appl. Environ. Microbiol. 2017, 83, e00947-17. [Google Scholar] [CrossRef] [PubMed]
- Kato, F.; Sugai, M. A simple method of markerless gene deletion in Staphylococcus aureus. J. Microbiol. Methods 2011, 87, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Oh, M.H.; Lee, J.C.; Kim, J.; Choi, C.H.; Han, K. Simple Method for Markerless Gene Deletion in Multidrug-Resistant Acinetobacter baumannii. Appl. Environ. Microbiol. 2015, 81, 3357–3368. [Google Scholar] [CrossRef] [PubMed]
- Junges, R.; Khan, R.; Tovpeko, Y.; Åmdal, H.A.; Petersen, F.C.; Morrison, D.A. Markerless Genome Editing in Competent Streptococci. Methods Mol. Biol. 2017, 1537, 233–247. [Google Scholar] [PubMed]
- Van Dam, V.; Bos, M.P. Generating knock-out and complementation strains of Neisseria meningitidis. Methods Mol. Biol. 2012, 799, 55–72. [Google Scholar] [CrossRef] [PubMed]
- Trebosc, V.; Gartenmann, S.; Royet, K.; Manfredi, P.; Tötzl, M.; Schellhorn, B.; Pieren, M.; Tigges, M.; Lociuro, S.; Sennhenn, P.C.; et al. A Novel Genome-Editing Platform for Drug-Resistant Acinetobacter baumannii Reveals an AdeR-Unrelated Tigecycline Resistance Mechanism. Antimicrob. Agents Chemother. 2016, 60, 7263–7271. [Google Scholar] [PubMed]
- White, A.P.; Allen-Vercoe, E.; Jones, B.W.; DeVinney, R.; Kay, W.W.; Surette, M.G. An efficient system for markerless gene replacement applicable in a wide variety of enterobacterial species. Can. J. Microbiol. 2007, 53, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Geng, S.; Tian, Q.; An, S.; Pan, Z.; Chen, X.; Jiao, X. High-Efficiency, Two-Step Scarless-Markerless Genome Genetic Modification in Salmonella enterica. Curr. Microbiol. 2016, 72, 700–706. [Google Scholar] [CrossRef] [PubMed]
- Pyne, M.E.; Bruder, M.R.; Moo-Young, M.; Chung, D.A.; Chou, C.P. Harnessing heterologous and endogenous CRISPR-Cas machineries for efficient markerless genome editing in Clostridium. Sci. Rep. 2016, 6, 25666. [Google Scholar] [CrossRef] [PubMed]
- Plaut, R.D.; Stibitz, S. Improvements to a Markerless Allelic Exchange System for Bacillus anthracis. PLoS ONE 2015, 10, e0142758. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.J.; Thurlow, C.M.; Sun, D.; Nasrin, S.; Liles, M.R. Genome modifications and cloning using a conjugally transferable recombineering system. Biotechnol. Rep. 2015, 8, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, Y.; Dai, K.; Wen, X.; Wu, R.; Huang, X.; Jin, J.; Xu, K.; Yan, Q.; Huang, Y.; et al. Establishment of a Successive Markerless Mutation System in Haemophilus parasuis through Natural Transformation. PLoS ONE 2015, 10, e0127393. [Google Scholar] [CrossRef] [PubMed]
- Gómez, E.; Álvarez, B.; Duchaud, E.; Guijarro, J.A. Development of a markerless deletion system for the fish-pathogenic bacterium Flavobacterium psychrophilum. PLoS ONE 2015, 10, e0117969. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Yang, D.; Wang, Y.; Geng, H.; He, X.; Liu, H. Development of a markerless gene deletion system for Streptococcus zooepidemicus: Functional characterization of hyaluronan synthase gene. Appl. Microbiol. Biotechnol. 2013, 97, 8629–8636. [Google Scholar] [CrossRef] [PubMed]
- Horzempa, J.; Shanks, R.M.; Brown, M.J.; Russo, B.C.; O’Dee, D.M.; Nau, G.J. Utilization of an unstable plasmid and the I-SceI endonuclease to generate routine markerless deletion mutants in Francisella tularensis. J. Microbiol. Methods 2010, 80, 106–108. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Wang, S.; Curtiss, R. Highly efficient method for introducing successive multiple scarless gene deletions and markerless gene insertions into the Yersinia pestis chromosome. Appl. Environ. Microbiol. 2008, 74, 4241–4245. [Google Scholar] [CrossRef] [PubMed]
- Kristich, C.J.; Chandler, J.R.; Dunny, G.M. Development of a host-genotype-independent counterselectable marker and a high-frequency conjugative delivery system and their use in genetic analysis of Enterococcus faecalis. Plasmid 2007, 57, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Górski, A.; Ważna, E.; Weber-Dąbrowska, B.; Dąbrowska, K.; Świtała-Jeleń, K.; Międzybrodzki, R. Bacteriophage translocation. FEMS Immunol. Med. Microbiol. 2006, 46, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Barr, J.J. A bacteriophage journey through the human body. Immunol. Rev. 2017, 279, 106–122. [Google Scholar] [CrossRef] [PubMed]
- Górski, A.; Dąbrowska, K.; Międzybrodzki, R.; Weber-Dąbrowska, B.; Łusiak-Szelachowska, M.; Jończyk-Matysiak, E.; Borysowski, J. Phages and immunomodulation. Future Microbiol. 2017. [Google Scholar] [CrossRef]
- Górski, A.; Jończyk-Matysiak, E.; Łusiak-Szelachowska, M.; Międzybrodzki, R.; Weber-Dąbrowska, B.; Borysowski, J. Bacteriophages targeting intestinal epithelial cells: A potential novel form of immunotherapy. Cell. Mol. Life Sci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Górski, A.; Jończyk-Matysiak, E.; Łusiak-Szelachoeska, M.; Weber-Dąbrowska, B.; Międzybrodzki, R.; Borysowski, J. Therapeutic potential of phages in autoimmune liver diseases. Clin. Exp. Immunol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Górski, A.; Jończyk-Matysiak, E.; Łusiak-Szelachowska, M.; Międzybrodzki, R.; Weber-Dąbrowska, B.; Borysowski, J. Phage therapy in allergic disorders? Exp. Biol. Med. 2018, 1. [Google Scholar] [CrossRef] [PubMed]
- Górski, A.; Jończyk-Matysiak, E.; Łusiak-Szelachowska, M.; Międzybrodzki, R.; Weber-Dąbrowska, B.; Borysowski, J. The potential of phage therapy in sepsis. Front. Immunol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Truog, R.D. The UK sets limits on experimental treatments. JAMA 2017, 318, 1001–1002. [Google Scholar] [CrossRef] [PubMed]
- Górski, A.; Jończyk-Matysiak, E.; Międzybrodzki, R.; Weber-Dąbrowska, B.; Łusiak-Szelachowska, M.; Bagińska, N.; Borysowski, J.; Lobocka, M.B.; Węgrzyn, A.; Wegrzyn, G. Phage therapy: Beyond the antibacterial action. Front. Med. 2018. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Górski, A.; Międzybrodzki, R.; Łobocka, M.; Głowacka-Rutkowska, A.; Bednarek, A.; Borysowski, J.; Jończyk-Matysiak, E.; Łusiak-Szelachowska, M.; Weber-Dąbrowska, B.; Bagińska, N.; et al. Phage Therapy: What Have We Learned? Viruses 2018, 10, 288. https://doi.org/10.3390/v10060288
Górski A, Międzybrodzki R, Łobocka M, Głowacka-Rutkowska A, Bednarek A, Borysowski J, Jończyk-Matysiak E, Łusiak-Szelachowska M, Weber-Dąbrowska B, Bagińska N, et al. Phage Therapy: What Have We Learned? Viruses. 2018; 10(6):288. https://doi.org/10.3390/v10060288
Chicago/Turabian StyleGórski, Andrzej, Ryszard Międzybrodzki, Małgorzata Łobocka, Aleksandra Głowacka-Rutkowska, Agnieszka Bednarek, Jan Borysowski, Ewa Jończyk-Matysiak, Marzanna Łusiak-Szelachowska, Beata Weber-Dąbrowska, Natalia Bagińska, and et al. 2018. "Phage Therapy: What Have We Learned?" Viruses 10, no. 6: 288. https://doi.org/10.3390/v10060288
APA StyleGórski, A., Międzybrodzki, R., Łobocka, M., Głowacka-Rutkowska, A., Bednarek, A., Borysowski, J., Jończyk-Matysiak, E., Łusiak-Szelachowska, M., Weber-Dąbrowska, B., Bagińska, N., Letkiewicz, S., Dąbrowska, K., & Scheres, J. (2018). Phage Therapy: What Have We Learned? Viruses, 10(6), 288. https://doi.org/10.3390/v10060288