Functional Scanning of Apple Geminivirus Proteins as Symptom Determinants and Suppressors of Posttranscriptional Gene Silencing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Construction of Plasmids
2.2. Agroinfiltration
2.3. Fluorescence Observation
2.4. H2O2 Detection in Plants
2.5. PTGS Assay
2.6. RNA and Protein Analyses
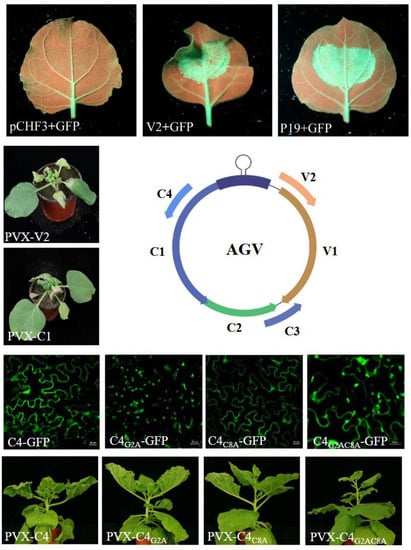
3. Results
3.1. Subcellular Localization
3.2. AGV V2 is a Suppressor of PTGS
3.3. V2 and C1 Potentially Enhance the Pathogenicity of PVX and C4 is a Putative Symptom Determinant
3.4. The N-terminal Acylated Sites of C4 are Associated with its Function as Symptom Determinant
4. Discussion
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Zerbini, F.M.; Briddon, R.W.; Idris, A.; Martin, D.P.; Moriones, E.; Navas-Castillo, J.; Rivera-Bustamante, R.; Roumagnac, P.; Varsani, A. ICTV virus taxonomy profile: Geminiviridae. J. Gen. Virol. 2017, 98, 131–133. [Google Scholar] [CrossRef] [PubMed]
- Mansoor, S.; Zafar, Y.; Briddon, R.W. Geminivirus disease complexes: the threat is spreading. Trends Plant Sci. 2006, 11, 209–212. [Google Scholar] [CrossRef] [PubMed]
- Varsani, A.; Roumagnac, P.; Fuchs, M.; Navas-Castillo, J.; Moriones, E.; Idris, A.; Briddon, R.W.; Rivera-Bustamante, R.; Murilo, Z.F.; Martin, D.P. Capulavirus and Grablovirus: Two new genera in the family Geminiviridae. Arch. Virol. 2017, 162, 1819–1831. [Google Scholar] [CrossRef] [PubMed]
- Polston, J.E.; Londoño, M.A.; Capobianco, H. The complete genome sequence of new world jatropha mosaic virus. Arch. Virol. 2014, 159, 3131–3136. [Google Scholar] [CrossRef] [PubMed]
- Narayana, D.S.A.; Shankarappa, K.S.; Govindappa, M.R.; Prameela, H.A.; Rao, M.R.G.; Rangaswamy, K.T. Natural occurrence of jatropha mosaic virus disease in India. Curr. Sci. 2006, 91, 584–586. [Google Scholar]
- Srivastava, A.; Kumar, S.; Jaidi, M.; Raj, S.K. Molecular characterization of a new begomovirus associated with leaf yellow mosaic disease of Jatropha curcas in India. Arch. Virol. 2015, 160, 1359–1362. [Google Scholar] [CrossRef] [PubMed]
- Rwahnih, M.A.; Dave, A.; Anderson, M.M.; Rowhani, A.; Uyemoto, J.K.; Sudarshana, M.R. Association of a DNA virus with grapevines affected by red blotch disease in California. Phytopathology 2013, 103, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- Rwahnih, M.A.; Rowhani, A.; Golino, D.A.; Islas, C.M.; Preece, J.E.; Sudarshana, M.R. Detection and genetic diversity of grapevine red blotch-associated virus isolates in table grape accessions in the National Clonal Germplasm Repository in California. Can. J. Plant Pathol. 2015, 37, 130–135. [Google Scholar] [CrossRef]
- Sudarshana, M.R.; Perry, K.L.; Fuchs, M.F. Grapevine red blotch-associated virus, an emerging threat to the grapevine industry. Phytopathology 2015, 105, 1026–1032. [Google Scholar] [CrossRef] [PubMed]
- Loconsole, G.; Saldarelli, P.; Doddapaneni, H.; Savino, V.; Martelli, G.P.; Saponari, M. Identification of a single-stranded DNA virus associated with citrus chlorotic dwarf disease, a new member in the family Geminiviridae. Virology 2012, 432, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Navarro, B.; Zhang, Z.; Lu, M.; Zhou, X.; Chi, S.; Di, S.F.; Li, S. Identification and molecular characterization of a novel monopartite geminivirus associated with mulberry mosaic dwarf disease. J. Gen. Virol. 2015, 96, 2421–2434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, P.; Navarro, B.; Zhang, Z.; Wang, H.; Lu, M.; Xiao, H.; Wu, Q.; Zhou, X.; Di, S.F.; Li, S. Identification and characterization of a novel geminivirus with a monopartite genome infecting apple trees. J. Gen. Virol. 2015, 96, 2411–2420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al, R.M.; Alabi, O.J.; Westrick, N.M.; Golino, D.; Rowhani, A. Description of a novel monopartite geminivirus and its defective subviral genome in grapevine. Phytopathology 2017, 107, 240–251. [Google Scholar]
- Sharma, P.; Ikegami, M. Tomato leaf curl Java virus V2 protein is a determinant of virulence, hypersensitive response and suppression of posttranscriptional gene silencing. Virology 2010, 396, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Ahlquist, P. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat. Rev. Microbiol. 2006, 4, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Fondong, V.N. Geminivirus protein structure and function. Mol. Plant Pathol. 2013, 14, 635–649. [Google Scholar] [CrossRef] [PubMed]
- Rojas, M.R.; Jiang, H.; Salati, R.; Xoconostle-Cázares, B.; Sudarshana, M.R.; Lucas, W.J.; Gilbertson, R.L. Functional analysis of proteins involved in movement of the monopartite begomovirus, tomato yellow leaf curl virus. Virology 2001, 291, 110–125. [Google Scholar]
- Sharma, P.; Ikegami, M. Characterization of signals that dictate nuclear/nucleolar and cytoplasmic shuttling of the capsid protein of tomato leaf curl Java virus associated with DNA beta satellite. Virus Res. 2009, 144, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Ding, S.W. Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 2006, 60, 503–531. [Google Scholar] [CrossRef] [PubMed]
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013, 11, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Vanitharani, R.; Chellappan, P.; Pita, J.S.; Fauquet, C.M. Differential roles of AC2 and AC4 of cassava geminiviruses in mediating synergism and suppression of posttranscriptional gene silencing. J. Virol. 2004, 78, 9487–9498. [Google Scholar] [CrossRef] [PubMed]
- Gopal, P.; Pravin, K.P.; Sinilal, B.; Jose, J.; Kasin, Y.A.; Usha, R. Differential roles of C4 and betaC1 in mediating suppression of post-transcriptional gene silencing: evidence for transactivation by the C2 of bhendi yellow vein mosaic virus, a monopartite begomovirus. Virus Res. 2007, 123, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Nawaz-Ul-Rehman, M.S.; Nahid, N.; Mansoor, S.; Briddon, R.W.; Fauquet, C.M. Post-transcriptional gene silencing suppressor activity of two non-pathogenic alphasatellites associated with a begomovirus. Virology 2010, 405, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Luna, A.P.; Rodrígueznegrete, E.A.; Morilla, G.; Wang, L.; Lozanodurán, R.; Castillo, A.G.; Bejarano, E.R. V2 from a curtovirus is a suppressor of post-transcriptional gene silencing. J. Gen. Virol. 2017, 98, 2607–2614. [Google Scholar] [CrossRef] [PubMed]
- Glick, E.; Zrachya, A.; Levy, Y.; Mett, A.; Gidoni, D.; Belausov, E.; Citovsky, V.; Gafni, Y. Interaction with host SGS3 is required for suppression of RNA silencing by tomato yellow leaf curl virus V2 protein. Proc. Natl. Acad. Sci. USA 2008, 105, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Zrachya, A.; Glick, E.; Levy, Y.; Arazi, T.; Citovsky, V.; Gafni, Y. Suppressor of RNA silencing encoded by tomato yellow leaf curl virus-Israel. Virology 2007, 358, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, R.; Doudna, J.A. DsRNA with 5′ overhangs contributes to endogenous and antiviral RNA silencing pathways in plants. EMBO J. 2009, 28, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Michael, D.C.; Katherine, M.L. Toward understanding the molecular mechanism of a geminivirus C4 protein. Plant Signal Behav. 2015, 10, e1109758. [Google Scholar]
- Piroux, N.; Saunders, K.; Page, A.; Stanley, J. Geminivirus pathogenicity protein C4 interacts with Arabidopsis thaliana shaggy-related protein kinase AtSKeta, a component of the brassinosteroid signalling pathway. Virology 2007, 362, 428–440. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.; Chen, H.; Teng, K.; Zhao, Q.; Zhang, Z.; Li, Y.; Liang, L.; Xia, R.; Wu, Y.; Guo, H. RKP, a RING finger E3 ligase induced by BSCTV C4 protein, affects geminivirus infection by regulation of the plant cell cycle. Plant J. 2009, 57, 905–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bi, H.; Fan, W.; Zhang, P. C4 protein of sweet potato leaf curl virus regulates brassinosteroid signaling pathway through interaction with AtBIN2 and affects male fertility in Arabidopsis. Front. Plant Sci. 2017, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Yang, X.; Huang, C.; Zhang, X.; Zhou, X. Tomato leaf curl Yunnan virus-encoded C4 induces cell division through enhancing stability of Cyclin D 1.1 via impairing NbSKη -mediated phosphorylation in Nicotiana benthamiana. PLoS Pathog. 2018, 14, e1006789. [Google Scholar] [CrossRef] [PubMed]
- Millslujan, K.; Andrews, D.L.; Chou, C.; Deom, C.M. The roles of phosphorylation and SHAGGY-like protein kinases in geminivirus C4 protein induced hyperplasia. Plos One 2015, 10, e0122356. [Google Scholar]
- Fondong, V.N.; Reddy, R.V.; Lu, C.; Hankoua, B.; Felton, C.; Czymmek, K.; Achenjang, F. The consensus N-myristoylation motif of a geminivirus AC4 protein is required for membrane binding and pathogenicity. Mol. Plant Microbe Interact. 2007, 20, 380–391. [Google Scholar] [CrossRef] [PubMed]






© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhan, B.; Zhao, W.; Li, S.; Yang, X.; Zhou, X. Functional Scanning of Apple Geminivirus Proteins as Symptom Determinants and Suppressors of Posttranscriptional Gene Silencing. Viruses 2018, 10, 488. https://doi.org/10.3390/v10090488
Zhan B, Zhao W, Li S, Yang X, Zhou X. Functional Scanning of Apple Geminivirus Proteins as Symptom Determinants and Suppressors of Posttranscriptional Gene Silencing. Viruses. 2018; 10(9):488. https://doi.org/10.3390/v10090488
Chicago/Turabian StyleZhan, Binhui, Wenyang Zhao, Shifang Li, Xiuling Yang, and Xueping Zhou. 2018. "Functional Scanning of Apple Geminivirus Proteins as Symptom Determinants and Suppressors of Posttranscriptional Gene Silencing" Viruses 10, no. 9: 488. https://doi.org/10.3390/v10090488
APA StyleZhan, B., Zhao, W., Li, S., Yang, X., & Zhou, X. (2018). Functional Scanning of Apple Geminivirus Proteins as Symptom Determinants and Suppressors of Posttranscriptional Gene Silencing. Viruses, 10(9), 488. https://doi.org/10.3390/v10090488