Algal Viruses: The (Atomic) Shape of Things to Come
Abstract
:1. Introduction
Early Virus Structural Studies
2. Electron Microscopy
2.1. Cryoelectron Microscopy
2.2. Electron Microscopy Viral ‘Dynamics’
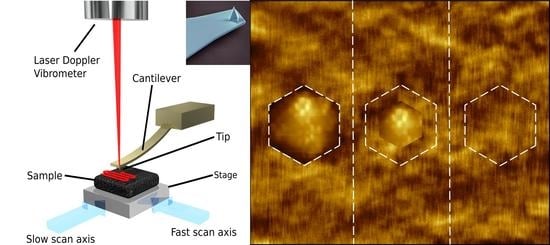
3. Atomic Force Microscopy
3.1. PBCV-1 Atomic Force Microscopy
3.2. Atomic Force Microscopy Viral ‘Dynamics’
3.3. Improving AFM Speeds
3.4. Force-Distance Curve-Based Atomic Force Microscopy
3.5. Subsurface Atomic Force Microscopy
4. Conclusions and Perspective
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bergh, O.; BOrsheim, K.Y.; Bratbak, G.; Heldal, M. High abundance of viruses found in aquatic environments. Nature 1989, 340, 467–468. [Google Scholar] [CrossRef] [PubMed]
- Breitbart, M.; Thompson, L.; Suttle, C.; Sullivan, M. Exploring the Vast Diversity of Marine Viruses. Oceanography 2007, 20, 135–139. [Google Scholar] [CrossRef] [Green Version]
- Guidi, L.; Chaffron, S.; Bittner, L.; Eveillard, D.; Larhlimi, A.; Roux, S.; Darzi, Y.; Audic, S.; Berline, L.; Brum, J.R.; et al. Plankton networks driving carbon export in the oligotrophic ocean. Nature 2016, 532, 465–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danovaro, R.; Corinaldesi, C.; Dell’Anno, A.; Fuhrman, J.A.; Middelburg, J.J.; Noble, R.T.; Suttle, C.A. Marine viruses and global climate change. FEMS Microbiol. Rev. 2011, 35, 993–1034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crick, F.H.C.; Watson, J.D. Structure of Small Viruses. Nature 1956, 177, 473–475. [Google Scholar] [CrossRef] [PubMed]
- Caspar, D.L.D. Structure of small viruses-tomato bushy stunt virus. Nature 1956, 177, 79–81. [Google Scholar] [CrossRef]
- Kaesberg, P. Structure of Small “Spherical” Viruses. Science 1956, 124, 626–629. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.C.; Smith, K.M. The polyhedral form of the Tipula iridescent virus. Biochim. Biophys. Acta 1958, 28, 464–469. [Google Scholar] [CrossRef]
- Steinmann, E. An Eletron Microscope Study of the Structure of Sericesthis Iridescent Virus. J. Gen. Virol. 1969, 5, 123–134. [Google Scholar]
- Vidaver, A.K.; Koski, R.K.; Van Etten, J.L. Bacteriophage phi6: A Lipid-Containing Virus of Pseudomonas phaseolicola. J. Virol. 1973, 11, 799–805. [Google Scholar] [PubMed]
- Dodds, J.A. Viruses of marine algae. Experientia 1979, 35, 440–442. [Google Scholar] [CrossRef]
- Van Etten, J.L.; Burbank, D.E.; Kuczmarski, D.; Meints, R.H. Virus Infection of Culturable Chlorella-Like Algae and Development of a Plaque Assay. Science 1983, 219, 994–996. [Google Scholar] [CrossRef] [PubMed]
- Von Ardenne, M.B. Die praktische Ausführung der Elektronensonden-Mikroskope. In Elektronen-Übermikroskopie; Springer: Berlin/Heidelberg, Germany, 1940. [Google Scholar]
- Mulvey, T. Origins and Historical Development of the Electron Microscope. Br. J. Appl. Phys. 1962, 13, 197–207. [Google Scholar] [CrossRef]
- Oatley, C.W. The early history of the scanning electron microscope. J. Appl. Phys. 1982, 53, R1–R13. [Google Scholar] [CrossRef]
- Meints, R.H.; Lee, K.; Burbank, D.E.; van Etten, J.L. Infection of a chlorella-like alga with the virus, PBCV-1: Ultrastructural studies. Virology 1984, 346, 341–346. [Google Scholar] [CrossRef]
- Skrdla, M.P.; Burbank, D.E.; Xia, Y.; Meints, R.H.; van Etten, J.L. Structural proteins and lipids in a virus, PBCV-1, which replicates in a Chlorella-like alga. Virology 1984, 135, 308–315. [Google Scholar] [CrossRef]
- Gazzarrini, S.; Severino, M.; Lombardi, M.; Morandi, M.; DiFrancesco, D.; Van Etten, J.L.; Thiel, G.; Moroni, A. The viral potassium channel Kcv: Structural and functional features. FEBS Lett. 2003, 552, 12–16. [Google Scholar] [CrossRef]
- Adrian, M.; Dubochet, J.; Lepault, J.; McDowall, A.W. Cryo-electron microscopy of viruses. Nature 1984, 308, 32–36. [Google Scholar] [CrossRef] [PubMed]
- The Nobel Prize in Chemistry 2017-Scientific Background: The Development of Cryo-Electron Microscopy. Available online: http://www.nobelprize.org/nobel_prizes/chemistry/laureates/2017/advanced.html (accessed on 29 July 2018).
- Yan, X.; Olson, N.H.; Etten, J.L.; Van Baker, T.S. Cryo-electron microscopy and image reconstruction of PBCV-1, an algal virus with T = 169 lattice symmetry. Electron. Microsc. 1998, 1, 775–776. [Google Scholar]
- Caspar, D.L.; Klug, A. Physical principles in the construction of regular viruses. Cold Spring Harb. Symp. Quant. Biol. 1962, 27, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xiang, Y.; Dunigan, D.D.; Klose, T.; Chipman, P.R.; Van Etten, J.L.; Rossmann, M.G. Three-dimensional structure and function of the Paramecium bursaria chlorella virus capsid. Proc. Natl. Acad. Sci. USA 2011, 108, 14837–14842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bushby, A.J.; P’Ng, K.M.Y.; Young, R.D.; Pinali, C.; Knupp, C.; Quantock, A.J. Imaging three-dimensional tissue architectures by focused ion beam scanning electron microscopy. Nat. Protoc. 2011, 6, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Aoyama, K.; Takagi, T.; Hirase, A.; Miyazawa, A. STEM tomography for thick biological specimens. Ultramicroscopy 2008, 109, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.Y.; Johnson, J.E. Viral life cycles captured in three-dimensions with electron microscopy tomography. Curr. Opin. Virol. 2011, 1, 125–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yashchenko, V.V.; Gavrilova, O.V.; Rautian, M.S.; Jakobsen, K.S. Association of Paramecium bursaria Chlorella viruses with Paramecium bursaria cells: Ultrastructural studies. Eur. J. Protistol. 2012, 48, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Thiel, G.; Dunigan, D.; Etten, J.L.; Van Etten, J.L. Van Progress in botany. Nature 1962, 194, 1023. [Google Scholar]
- Meints, R.H.; Lee, K.; Van Etten, J.L. Assembly site of the virus PBCV-1 in a Chlorella-like green alga: Ultrastructural studies. Virology 1986, 154, 240–245. [Google Scholar] [CrossRef]
- Milrot, E.; Mutsafi, Y.; Fridmann-Sirkis, Y.; Shimoni, E.; Rechav, K.; Gurnon, J.R.; Van Etten, J.L.; Minsky, A. Virus-host interactions: Insights from the replication cycle of the large Paramecium bursaria chlorella virus. Cell Microbiol. 2016, 18, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Murata, K.; Zhang, Q.; Galaz-Montoya, J.M.; Fu, C.; Coleman, M.L.; Osburne, M.S.; Schmid, M.F.; Sullivan, M.B.; Chisholm, S.W.; Chiu, W. Visualizing Adsorption of Cyanophage P-SSP7 onto Marine Prochlorococcus. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zeev-Ben-Mordehai, T.; Hagen, C.; Grunewald, K. A cool hybrid approach to the herpesvirus ‘life’ cycle. Curr. Opin. VIrol. 2014, 5, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Nandhagopal, N.; Simpson, A.A.; Gurnon, J.R.; Yan, X.; Baker, T.S.; Graves, M.V.; Van Etten, J.L.; Rossmann, M.G. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc. Natl. Acad. Sci. USA 2002, 99, 14758–14763. [Google Scholar] [CrossRef] [PubMed]
- De Castro, C.; Klose, T.; Speciale, I.; Lanzetta, R.; Molinaro, A.; Van Etten, J.L.; Rossmann, M.G. Structure of the chlorovirus PBCV-1 major capsid glycoprotein determined by combining crystallographic and carbohydrate molecular modeling approaches. Proc. Natl. Acad. Sci. USA 2017, 115, E44–E52. [Google Scholar] [CrossRef] [PubMed]
- Romani, G.; Piotrowski, A.; Hillmer, S.; Gurnon, J.; Van Etten, J.L.; Moroni, A.; Thiel, G.; Hertel, B. A virus-encoded potassium ion channel is a structural protein in the chlorovirus Paramecium bursaria chlorella virus 1 virion. J. Gen. Virol. 2013, 94, 2549–2556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thiel, G.; Greiner, T.; Dunigan, D.D.; Moroni, A.; Van Etten, J.L. Large dsDNA chloroviruses encode diverse membrane transport proteins. Virology 2015, 479–480, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Olson, N.H.; Van Etten, J.L.; Bergoin, M.; Rossmann, M.G.; Baker, T.S. Structure and assembly of large lipid-containing dsDNA viruses. Nat. Struct. Biol. 2000, 7, 101–103. [Google Scholar] [PubMed]
- Binnig, G.; Quate, C.F.; Gerber, C. Atomic Force Microscope. Phys. Rev. Lett. 1986, 56, 930–933. [Google Scholar] [CrossRef] [PubMed]
- Kasas, S.; Thomson, N.H.; Smith, B.L.; Hansma, P.K.; Miklossy, J.; Hansma, H.G. Biological applications of the AFM: From single molecules to organs. Int. J. Imaging Syst. Technol. 1997, 8, 151–161. [Google Scholar] [CrossRef]
- Häberle, W.; Hörber, J.K.H.; Ohnesorge, F.; Smith, D.P.E.; Binnig, G. In situ investigations of single living cells infected by viruses. Ultramicroscopy 1992, 42–44, 1161–1167. [Google Scholar] [CrossRef]
- Ohnesorge, F.M.; Hörber, J.K.H.; Häberle, W.; Czerny, C.P.; Smith, D.P.E.; Binnig, G. AFM review study on pox viruses and living cells. Biophys. J. 1997, 73, 2183–2194. [Google Scholar] [CrossRef] [Green Version]
- Payton, O.D.; Picco, L.; Scott, T.B. High-speed atomic force microscopy for materials science. Int. Mater. Rev. 2016, 6608, 1–22. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Sheng, S.T.; Shao, Z.F. Imaging biological structures with the cryo atomic force microscope. Biophys. J. 1996, 71, 2168–2176. [Google Scholar] [CrossRef] [Green Version]
- Proctor, L.M. Advances in the Study of Marine Viruses. Microsc. Res. Tech. 1997, 37, 136–161. [Google Scholar] [CrossRef]
- Malkin, A.J.; Kuznetsov, Y.G.; Lucas, R.W.; McPherson, A. Surface processes in the crystallization of turnip yellow mosaic virus visualized by atomic force microscopy. J. Struct. Biol. 1999, 127, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Q.; Inniss, D.; Kjoller, K.; Elings, V.B. Fractured polymer/silica fiber surface studied by tapping mode atomic force microscopy. Surf. Sci. 1993, 290, L688–L692. [Google Scholar] [CrossRef]
- Hansma, P.K.; Cleveland, J.P.; Radmacher, M.; Walters, D.A.; Hillner, P.E.; Bezanilla, M.; Fritz, M.; Vie, D.; Hansma, H.G.; Prater, C.B.; et al. Tapping mode atomic force microscopy in liquids. Appl. Phys. Lett. 1994, 64, 1738. [Google Scholar] [CrossRef]
- McPherson, A.; Kuznetsov, Y.G. Atomic force microscopy investigation of viruses. Methods Mol. Biol. 2001, 736, 171–195. [Google Scholar]
- Wagner, P. Immobilization strategies for biological scanning probe microscopy. FEBS Lett. 1998, 430, 112–115. [Google Scholar] [CrossRef]
- Kuznetsov, Y.G.; McPherson, A. Atomic Force Microscopy in Imaging of Viruses and Virus-Infected Cells. Microbiol. Mol. Biol. Rev. 2011, 75, 268–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuznetsov, Y.G.; Gurnon, J.R.; Van Etten, J.L.; McPherson, A. Atomic force microscopy investigation of a chlorella virus, PBCV-1. J. Struct. Biol. 2005, 149, 256–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuznetsov, Y.G.; Datta, S.; Kothari, N.H.; Greenwood, A.; Fan, H.; McPherson, A. Atomic force microscopy investigation of fibroblasts infected with wild-type and mutant murine leukemia virus (MuLV). Biophys. J. 2002, 83, 3665–3674. [Google Scholar] [CrossRef]
- Kuznetsov, Y.G.; Victoria, J.G.; Robinson, W.E.; Mcpherson, A. Atomic Force Microscopy Investigation of Human Immunodeficiency Virus (HIV) and HIV-Infected Lymphocytes. J. Virol. 2003, 77, 11896–11909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Low, A.; Datta, S.; Kuznetsov, Y.; Jahid, S.; Kothari, N.; McPherson, A.; Fan, H. Mutation in the glycosylated gag protein of murine leukemia virus results in reduced in vivo infectivity and a novel defect in viral budding or release. J. Virol. 2007, 81, 3685–3692. [Google Scholar] [CrossRef] [PubMed]
- Gladnikoff, M.; Rousso, I. Directly monitoring individual retrovirus budding events using atomic force microscopy. Biophys. J. 2008, 94, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Barrett, R.C.; Quate, C.F. High-speed, large-scale imaging with the atomic force microscope. J. Vac. Sci. Technol. B 1991, 9, 302. [Google Scholar] [CrossRef]
- Viani, M.B.; Schäffer, T.E.; Paloczi, G.T.; Pietrasanta, L.I.; Smith, B.L. Fast imaging and fast force spectroscopy of single biopolymers with a new atomic force microscope designed for small cantilevers. Rev. Sci. 1999, 70, 4300–4303. [Google Scholar] [CrossRef]
- Ando, T.; Kodera, N.; Naito, Y.; Kinoshita, T.; Furuta, K.; Toyoshima, Y.Y. A High-speed Atomic Force Microscope for Studying Biological Macromolecules in Action. ChemPhysChem 2003, 4, 1196–1202. [Google Scholar] [CrossRef] [PubMed]
- Humphris, A.D.L.; Miles, M.J.; Hobbs, J.K. A mechanical microscope: High-speed atomic force microscopy. Appl. Phys. Lett. 2005, 86, 034106. [Google Scholar] [CrossRef]
- Ando, T.; Uchihashi, T.; Fukuma, T. High-speed atomic force microscopy for nano-visualization of dynamic biomolecular processes. Prog. Surf. Sci. 2008, 83, 337–437. [Google Scholar] [CrossRef] [Green Version]
- Picco, L.M.; Dunton, P.G.; Ulcinas, A.; Engledew, D.J.; Hoshi, O.; Ushiki, T.; Miles, M.J. High-speed AFM of human chromosomes in liquid. Nanotechnology 2008, 19, 384018. [Google Scholar] [CrossRef] [PubMed]
- Picco, L.; Bozec, L.; Ulcinas, A.; Engledew, D.J.; Antognozzi, M.; Horton, M.; Miles, M.J. Breaking the speed limit with atomic force microscopy. Nanotechnology 2007, 18, 044030. [Google Scholar] [CrossRef]
- Kodera, N.; Yamamoto, D.; Ishikawa, R.; Ando, T. Video imaging of walking myosin V by high-speed atomic force microscopy. Nature 2010, 468, 72–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shibata, M.; Nishimasu, H.; Kodera, N.; Hirano, S.; Ando, T.; Uchihashi, T.; Nureki, O. Real-space and real-time dynamics of CRISPR-Cas9 visualized by high-speed atomic force microscopy. Nat. Commun. 2017, 8, 1430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ando, T.; Uchihashi, T.; Kodera, N.; Yamamoto, D.; Miyagi, A.; Taniguchi, M.; Yamashita, H. High-speed AFM and nano-visualization of biomolecular processes. Pflugers Arch. Eur. J. Physiol. 2008, 456, 211–225. [Google Scholar] [CrossRef] [PubMed]
- Ando, T. High-speed atomic force microscopy coming of age. Nanotechnology 2012, 23, 062001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ando, T.; Uchihashi, T.; Kodera, N. High-Speed AFM and Applications to Biomolecular Systems. Annu. Rev. Biophys. 2013, 42, 393–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ando, T. High-speed AFM imaging. Curr. Opin. Struct. Biol. 2014, 28, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Ando, T. Directly watching biomolecules in action by high-speed atomic force microscopy. Biophys. Rev. 2017, 9, 421–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ando, T. High-speed atomic force microscopy and its future prospects. Biophys. Rev. 2018, 10, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Hodel, A.W.; Leung, C.; Dudkina, N.V.; Saibil, H.R.; Hoogenboom, B.W. Atomic force microscopy of membrane pore formation by cholesterol dependent cytolysins. Curr. Opin. Struct. Biol. 2016, 39, 8–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, C.; Hodel, A.W.; Brennan, A.J.; Lukoyanova, N.; Tran, S.; House, C.M.; Kondos, S.C.; Whisstock, J.C.; Dunstone, M.A.; Trapani, J.A.; et al. Real-time visualization of perforin nanopore assembly. Nat. Nanotechnol. 2017, 12, 467–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sumino, A.; Uchihashi, T.; Oiki, S. Oriented Reconstitution of the Full-Length KcsA Potassium Channel in a Lipid Bilayer for AFM Imaging. J. Phys. Chem. Lett. 2017, 8, 785–973. [Google Scholar] [CrossRef] [PubMed]
- Mikheikin, A.; Olsen, A.; Leslie, K.; Mishra, B.; Gimzewski, J.K.; Reed, J. Atomic force microscopic detection enabling multiplexed low-cycle-number quantitative polymerase chain reaction for biomarker assays. Anal. Chem. 2014, 86, 6180–6183. [Google Scholar] [CrossRef] [PubMed]
- Mikheikin, A.; Olsen, A.; Picco, L.; Payton, O.; Mishra, B.; Gimzewski, J.K.; Reed, J. High-Speed Atomic Force Microscopy Revealing Contamination in DNA Purification Systems. Anal. Chem. 2016, 88, 2527–2532. [Google Scholar] [CrossRef] [PubMed]
- Mikheikin, A.; Olsen, A.; Leslie, K.; Russell-Pavier, F.; Yacoot, A.; Picco, L.; Payton, O.; Toor, A.; Chesney, A.; Gimzewski, J.K.; et al. DNA nanomapping using CRISPR-Cas9 as a programmable nanoparticle. Nat. Commun. 2017, 8, 1665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fritz, M.; Radmacher, M.; Gaub, H.E. Granula Motion and Membrane Spreading during Activation of Human Platelets Imaged by Atomic-Force Microscopy. Biophys. J. 1994, 66, 1328–1334. [Google Scholar] [CrossRef]
- Henderson, E. Imaging of living cells by atomic force microscopy. Prog. Surf. Sci. 1994, 46, 39–60. [Google Scholar] [CrossRef]
- Kasas, S.; Ikai, A. A method for anchoring round shaped cells for atomic force microscope imaging. Biophys. J. 1995, 68, 1678–1680. [Google Scholar] [CrossRef] [Green Version]
- Formosa, C.; Pillet, F.; Schiavone, M.; Duval, R.E.; Ressier, L.; Dague, E. Generation of living cell arrays for atomic force microscopy studies. Nat. Protoc. 2015, 10, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Gebeshuber, I.C.; Kindt, J.H.; Thomson, J.B.; Del Amo, Y.; Stachelbergers, H.; Brzezinski, M.A.; Stucky, G.D.; Morse, D.E.; Hansma, P.K. Atomic force microscopy study of living diatoms in ambient conditions. J. Microsc. 2003, 212, 292–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luís, A.T.; Hlúbiková, D.; Vaché, V.; Choquet, P.; Hoffmann, L.; Ector, L. Atomic force microscopy (AFM) application to diatom study: Review and perspectives. J. Appl. Phycol. 2017, 29, 2989–3001. [Google Scholar] [CrossRef]
- Callow, J.A.; Crawford, S.A.; Higgins, M.J.; Mulvaney, P.; Wetherbee, R. The application of atomic force microscopy to topographical studies and force measurements on the secreted adhesive of the green alga Enteromorpha. Planta 2000, 211, 641–647. [Google Scholar] [CrossRef] [PubMed]
- Shibata, M.; Uchihashi, T.; Ando, T.; Yasuda, R. Long-tip high-speed atomic force microscopy for nanometer-scale imaging in live cells. Sci. Rep. 2015, 5, 8724. [Google Scholar] [CrossRef] [PubMed]
- Berquand, A.; Xia, N.; Castner, D.G.; Clare, B.H.; Abbott, N.L.; Dupres, V.; Adriaensen, Y.; Dufrêne, Y.F. Antigen binding forces of single antilysozyme Fv fragments explored by atomic force microscopy. Langmuir 2005, 21, 5517–5523. [Google Scholar] [CrossRef] [PubMed]
- Dufrêne, Y.F. Using nanotechniques to explore microbial surfaces. Nat. Rev. Microbiol. 2004, 2, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Dufrêne, Y.F. Microbial Nanoscopy: Breakthroughs, Challenges, and Opportunities. ACS Nano 2017, 11, 19–22. [Google Scholar] [CrossRef] [PubMed]
- Alsteens, D.; Newton, R.; Schubert, R.; Martinez-Martin, D.; Delguste, M.; Roska, B.; Müller, D.J. Nanomechanical mapping of first binding steps of a virus to animal cells. Nat. Nanotechnol. 2016, 12, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Sieben, C.; Herrmann, A. Single virus force spectroscopy: The ties that bind. Nat. Nanotechnol. 2017, 12, 102–103. [Google Scholar] [CrossRef] [PubMed]
- Agarkova, I.; Hertel, B.; Zhang, X.; Lane, L.; Tchourbanov, A.; Dunigan, D.D.; Thiel, G.; Rossmann, M.G.; Van Etten, J.L. Dynamic attachment of Chlorovirus PBCV-1 to Chlorella variabilis. Virology 2014, 466–467, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Dufrêne, Y.F.; Ando, T.; Garcia, R.; Alsteens, D.; Martinez-Martin, D.; Engel, A.; Gerber, C.; Müller, D.J. Imaging modes of atomic force microscopy for application in molecular and cell biology. Nat. Nanotechnol. 2017, 12, 295–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cartagena, A.; Hernando-Pérez, M.; Carrascosa, J.L.; de Pablo, P.J.; Raman, A. Mapping in vitro local material properties of intact and disrupted virions at high resolution using multi-harmonic atomic force microscopy. Nanoscale 2013, 5, 4729. [Google Scholar] [CrossRef] [PubMed]
- Cartagena-Rivera, A.X.; Wang, W.H.; Geahlen, R.L.; Raman, A. Fast, multi-frequency, and quantitative nanomechanical mapping of live cells using the atomic force microscope. Sci. Rep. 2015, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- González-Domínguez, I.; Gutiérrez-Granados, S.; Cervera, L.; Gòdia, F.; Domingo, N. Identification of HIV-1-Based Virus-like Particles by Multifrequency Atomic Force Microscopy. Biophys. J. 2016, 111, 1173–1179. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, M.; Wuite, G.; Roos, W. Atomic force microscopy observation and characterization of single virions and virus-like particles by nano-indentation. Curr. Opin. Virol. 2016, 18, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Iwata, F.; Ohashi, Y.; Ishisaki, I.; Picco, L.M.; Ushiki, T. Development of nanomanipulator using a high-speed atomic force microscope coupled with a haptic device. Ultramicroscopy 2013, 133, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.; Park, I.; Lee, Y.; Kim, H.J.; Jung, J.H.; Lee, J.H.; Kim, Y.; Kim, J.H.; Park, J.W. Visualization and Quantification of MicroRNA in a Single Cell Using Atomic Force Microscopy. J. Am. Chem. Soc. 2016, 138, 11664–11671. [Google Scholar] [CrossRef] [PubMed]
- Plomp, M.; Rice, M.K.; Wagner, E.K.; Mcpherson, A.; Malkin, A.J. Rapid Visualization at High Resolution of Pathogens by Atomic Force Microscopy. Am. J. Pathol. 2002, 160, 1959–1966. [Google Scholar] [CrossRef]
- Malkin, A.J.; McPherson, A.; Gershon, P.D. Structure of intracellular mature vaccinia virus visualized by in situ atomic force microscopy. J. Virol. 2003, 77, 6332–6340. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, Y.G.; Victoria, J.G.; Low, A.; Robinson, W.E.; Fan, H.; McPherson, A. Atomic force microscopy imaging of retroviruses: Human immunodeficiency virus and murine leukemia virus. Scanning 2004, 26, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, Y.G.; McPherson, A. Atomic force microscopy investigation of Turnip Yellow Mosaic Virus capsid disruption and RNA extrusion. Virology 2006, 352, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, Y.G.; Daijogo, S.; Zhou, J.; Semler, B.L.; McPherson, A. Atomic force microscopy analysis of icosahedral virus RNA. J. Mol. Biol. 2005, 347, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, Y.G.; Dowell, J.J.; Gavira, J.A.; Ng, J.D.; McPherson, A. Biophysical and atomic force microscopy characterization of the RNA from satellite tobacco mosaic virus. Nucleic Acids Res. 2010, 38, 8284–8294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuznetsov, Y.G.; McPherson, A. Identification of DNA and RNA from retroviruses using ribonuclease A. Scanning 2006, 28, 278–281. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, Y.G.; Klose, T.; Rossmann, M.; McPherson, A. Morphogenesis of mimivirus and its viral factories: An atomic force microscopy study of infected cells. J. Virol. 2013, 87, 11200–11213. [Google Scholar] [CrossRef] [PubMed]
- Wulfmeyer, T.; Polzer, C.; Hiepler, G.; Hamacher, K.; Shoeman, R.; Dunigan, D.D.; van Etten, J.L.; Lolicato, M.; Moroni, A.; Thiel, G.; et al. Structural organization of DNA in chlorella viruses. PLoS ONE 2012, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.; Bestembayeva, A.; Thorogate, R.; Stinson, J.; Pyne, A.; Marcovich, C.; Yang, J.; Drechsler, U.; Despont, M.; Jankowski, T.; et al. Atomic force microscopy with nanoscale cantilevers resolves different structural conformations of the DNA double helix. Nano Lett. 2012, 12, 3846–3850. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.Y.-R.; Kudryashev, M.; Li, X.; Egelman, E.H.; Basler, M.; Cheng, Y.; Baker, D.; Dimaio, F. De novo Protein Structure Determination from Near-Atomic Resolution Cryo-EM Maps. Nat. Methods 2015, 12, 335–338. [Google Scholar] [CrossRef] [PubMed]
- Gross, L.; Wang, Z.L.; Ugarte, D.; Mohn, F.; Moll, N.; Heer, W.A.; Vincent, P.; Liljeroth, P.; Journet, C.; Meyer, G.; et al. The Chemical Structure of a Molecule Resolved by Atomic Force Microscopy. Science 2009, 325, 1110–1114. [Google Scholar] [CrossRef] [PubMed]
- Majzik, Z.; Pavliček, N.; Vilas-Varela, M.; Pérez, D.; Moll, N.; Guitián, E.; Meyer, G.; Peña, D.; Gross, L. Studying an antiaromatic polycyclic hydrocarbon adsorbed on different surfaces. Nat. Commun. 2018, 9, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Wastl, D.S.; Weymouth, A.J.; Giessibl, F.J. Atomically resolved graphitic surfaces in air by atomic force microscopy. ACS Nano 2014, 8, 5233–5239. [Google Scholar] [CrossRef] [PubMed]
- Wastl, D.S.; Judmann, M.; Weymouth, A.J.; Giessibl, F.J. Atomic resolution of calcium and oxygen sublattices of calcite in ambient conditions by atomic force microscopy using qPlus sensors with sapphire tips. ACS Nano 2015, 9, 3858–3865. [Google Scholar] [CrossRef] [PubMed]






© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Evans, C.T.; Payton, O.; Picco, L.; Allen, M.J. Algal Viruses: The (Atomic) Shape of Things to Come. Viruses 2018, 10, 490. https://doi.org/10.3390/v10090490
Evans CT, Payton O, Picco L, Allen MJ. Algal Viruses: The (Atomic) Shape of Things to Come. Viruses. 2018; 10(9):490. https://doi.org/10.3390/v10090490
Chicago/Turabian StyleEvans, Christopher T., Oliver Payton, Loren Picco, and Michael J. Allen. 2018. "Algal Viruses: The (Atomic) Shape of Things to Come" Viruses 10, no. 9: 490. https://doi.org/10.3390/v10090490
APA StyleEvans, C. T., Payton, O., Picco, L., & Allen, M. J. (2018). Algal Viruses: The (Atomic) Shape of Things to Come. Viruses, 10(9), 490. https://doi.org/10.3390/v10090490